Now that it has become evident that Late Repin (i.e. Yamnaya/Afanasevo) ancestry was associated with the migration of R1b-L23-rich Late Proto-Indo-Europeans from the steppe in the second half of the the 4th millennium BC, there’s still the question of how R1a-rich Uralic speakers of Corded Ware ancestry expanded , and how they spread their languages throughout North Eurasia.
Modern North Eurasians
I have been collecting information from the supplementary data of the latest papers on modern and ancient North Eurasian peoples, including Jeong et al. (2019), Saag et al. (2019), Sikora et al. (2018), or Flegontov et al. (2019), and I have tried to add up their information on ancestral components and their modern and historical distributions.
Fortunately, the current obsession with simplifying ancestry components into three or four general, atemporal groups, and the common use of the same ones across labs, make it very simple to merge data and map them.
Corded Ware ancestry
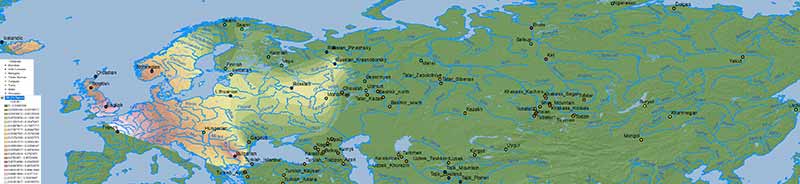
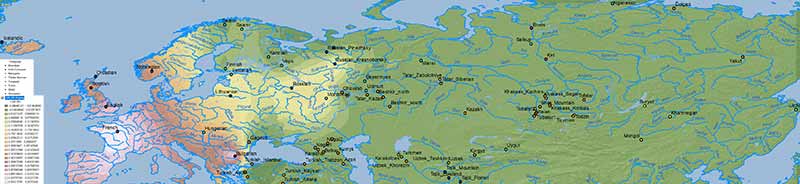
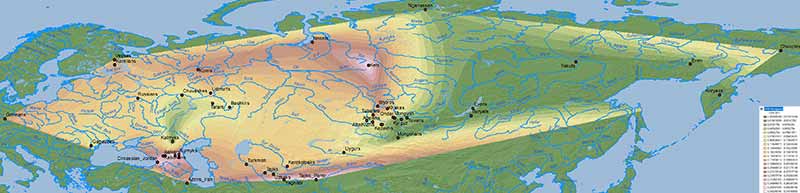
There is no doubt about the prevalent ancestry among Uralic-speaking peoples. A map isn’t needed to realize that, because ancient and modern data – like those recently summarized in Jeong et al. (2019) – prove it. But maps sure help visualize their intricate relationship better:
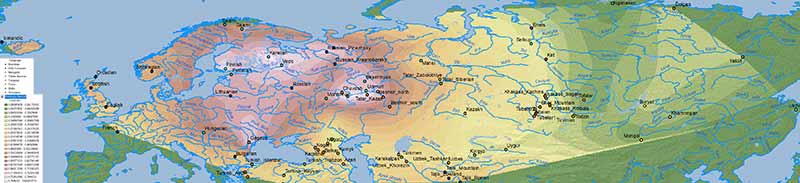
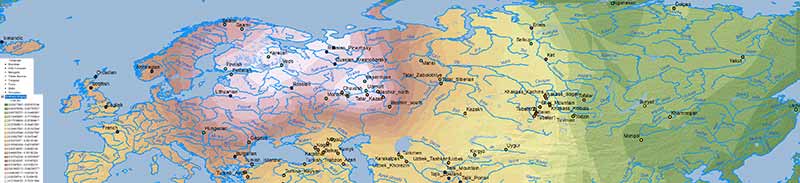
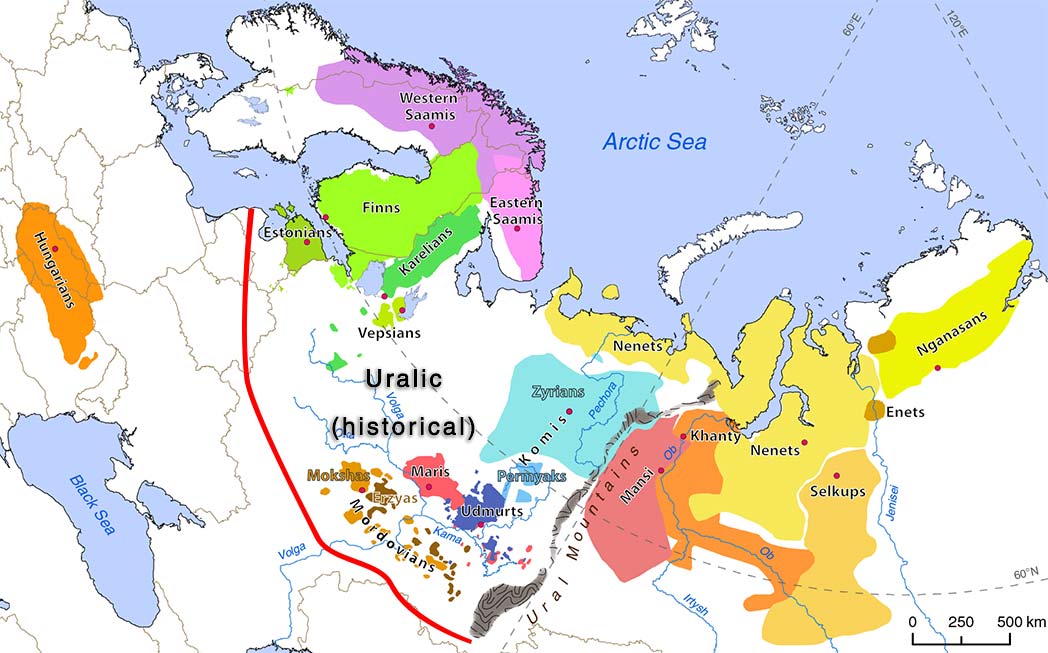
Interestingly, the regions with higher Corded Ware-related ancestry are in great part coincident with (pre)historical Finno-Ugric-speaking territories:
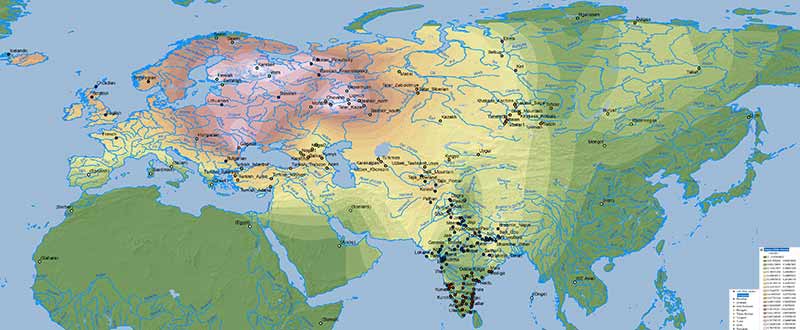
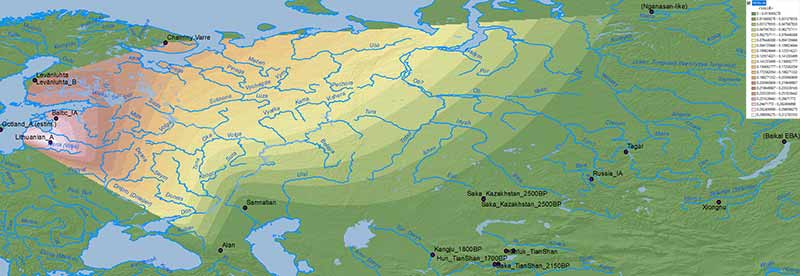
Edit (29/7/2019): Here is the full Steppe_MLBA ancestry map, including Steppe_MLBA (vs. Indus Periphery vs. Onge) in modern South Asian populations from Narasimhan et al. (2018), apart from the ‘Srubnaya component’ in North Eurasian populations. ‘Dummy’ variables (with 0% ancestry) have been included to the south and east of the map to avoid weird interpolations of Steppe_MLBA into Africa and East Asia.
Anatolia Neolithic ancestry
Also interesting are the patterns of non-CWC-related ancestry, in particular the apparent wedge created by expanding East Slavs, which seems to reflect the intrusion of central(-eastern) European ancestry into Finno-Permic territory.
NOTE. Read more on Balto-Slavic hydrotoponymy, on the cradle of Russians as a Finno-Permic hotspot, and about Pre-Slavic languages in North-West Russia.
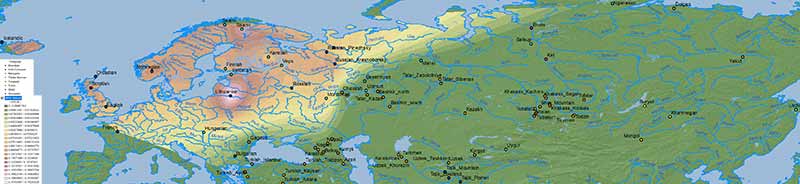
WHG ancestry
The cline(s) between WHG, EHG, ANE, Nganasan, and Baikal HG are also simplified when some of them excluded, in this case EHG, represented thus in part by WHG, and in part by more eastern ancestries (see below).
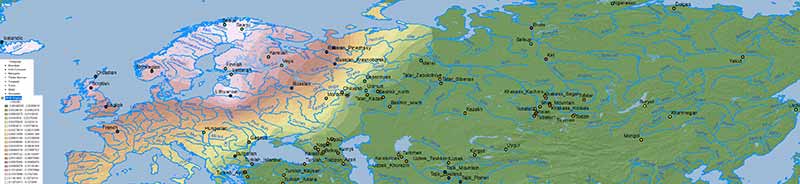
Arctic, Tundra or Forest-steppe?
Data on Nganasan-related vs. ANE vs. Baikal HG/Ulchi-related ancestry is difficult to map properly, because both ancestry components are usually reported as mutually exclusive, when they are in fact clearly related in an ancestral cline formed by different ancient North Eurasian populations from Siberia.
When it comes to ascertaining the origin of the multiple CWC-related clines among Uralic-speaking peoples, the question is thus how to properly distinguish the proportions of WHG-, EHG-, Nganasan-, ANE or BaikalHG-related ancestral components in North Eurasia, i.e. how did each dialectal group admix with regional groups which formed part of these clines east and west of the Urals.
The truth is, one ought to test specific ancient samples for each “Siberian” ancestry found in the different Uralic dialectal groups, but the simplistic “Siberian” label somehow gets a pass in many papers (see a recent example).
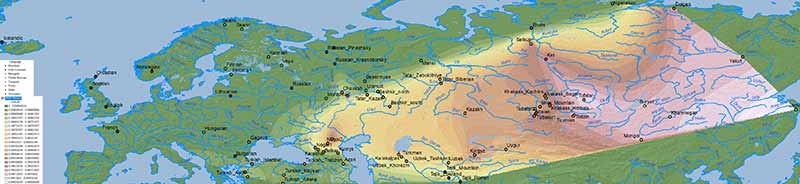
Below qpAdm results with best fits for Ulchi ancestry, Afontova Gora 3 ancestry, and Nganasan ancestry, but some populations show good fits for both and with similar proportions, so selecting one necessarily simplifies the distribution of both.
Ulchi ancestry
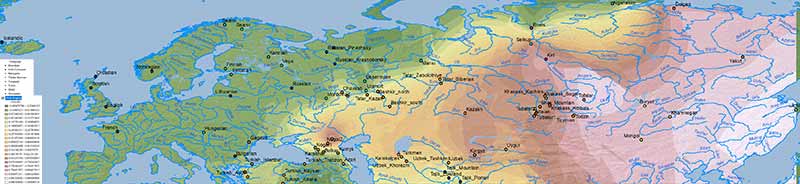
ANE ancestry
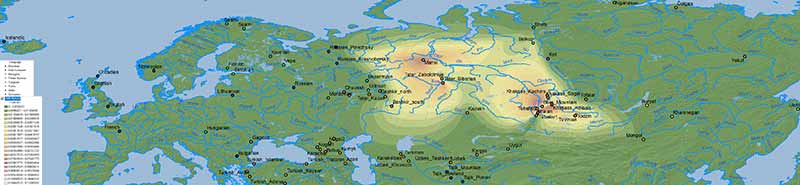
Nganasan ancestry
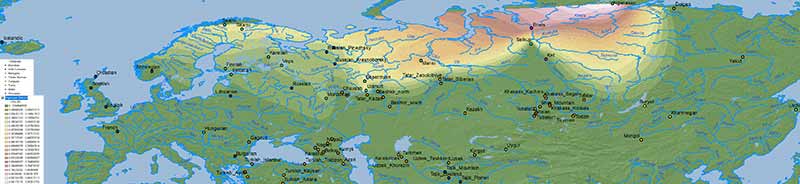
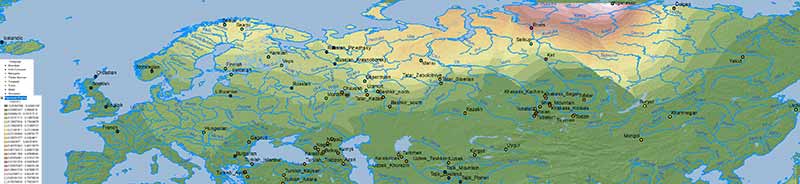
Iran Chalcolithic
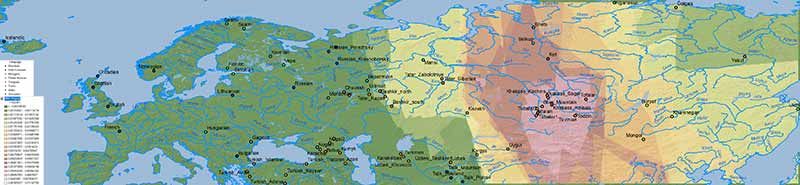
A simplistic Iran Chalcolithic-related ancestry is also seen in the Altaic cline(s) which (like Corded Ware ancestry) expanded from Central Asia into Europe – apart from its historical distribution south of the Caucasus:
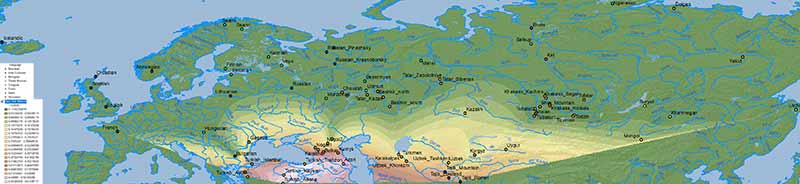
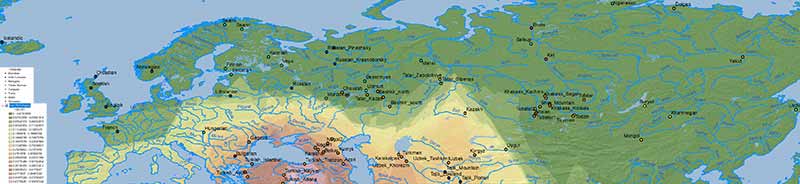
Other models
The first question I imagine some would like to know is: what about other models? Do they show the same results? Here is the simplistic combination of ancestry components published in Damgaard et al. (2018) for the same or similar populations:
NOTE. As you can see, their selection of EHG vs. WHG vs. Nganasan vs. Natufian vs. Clovis of is of little use, but corroborate the results from other papers, and show some interesting patterns in combination with those above.
EHG
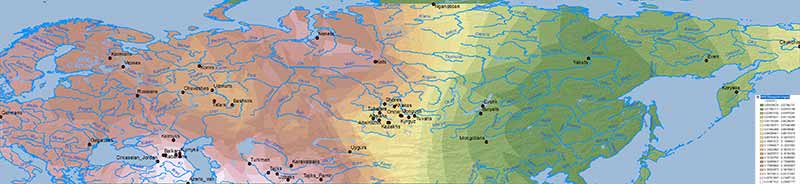
Natufian ancestry
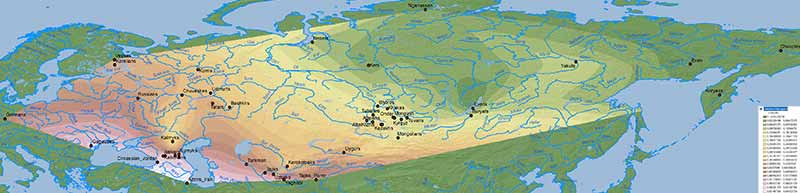
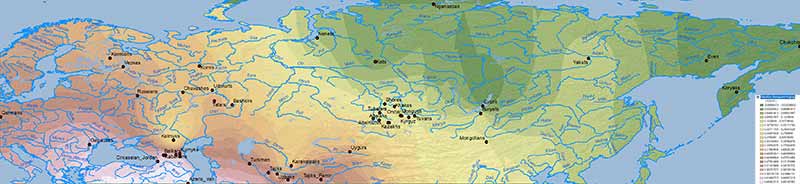
WHG ancestry
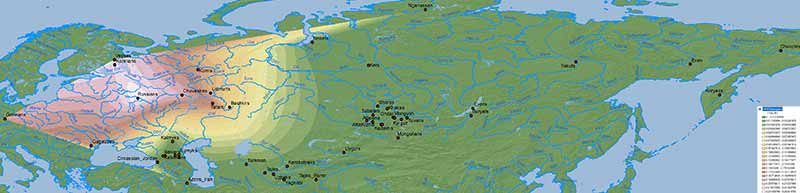
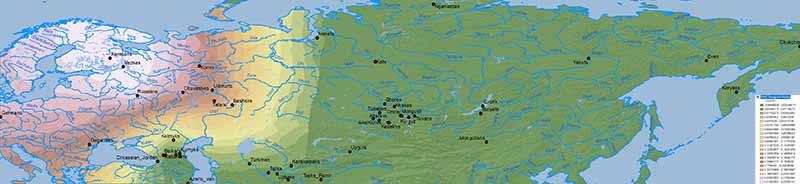
Baikal HG ancestry
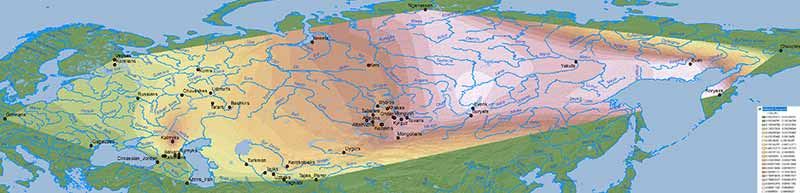
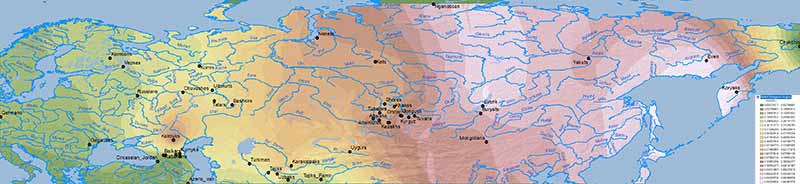
Ancient North Eurasians
Once the modern situation is clear, relevant questions are, for example, whether EHG-, WHG-, ANE, Nganasan-, and/or Baikal HG-related meta-populations expanded or became integrated into Uralic-speaking territories.
When did these admixture/migration events happen?
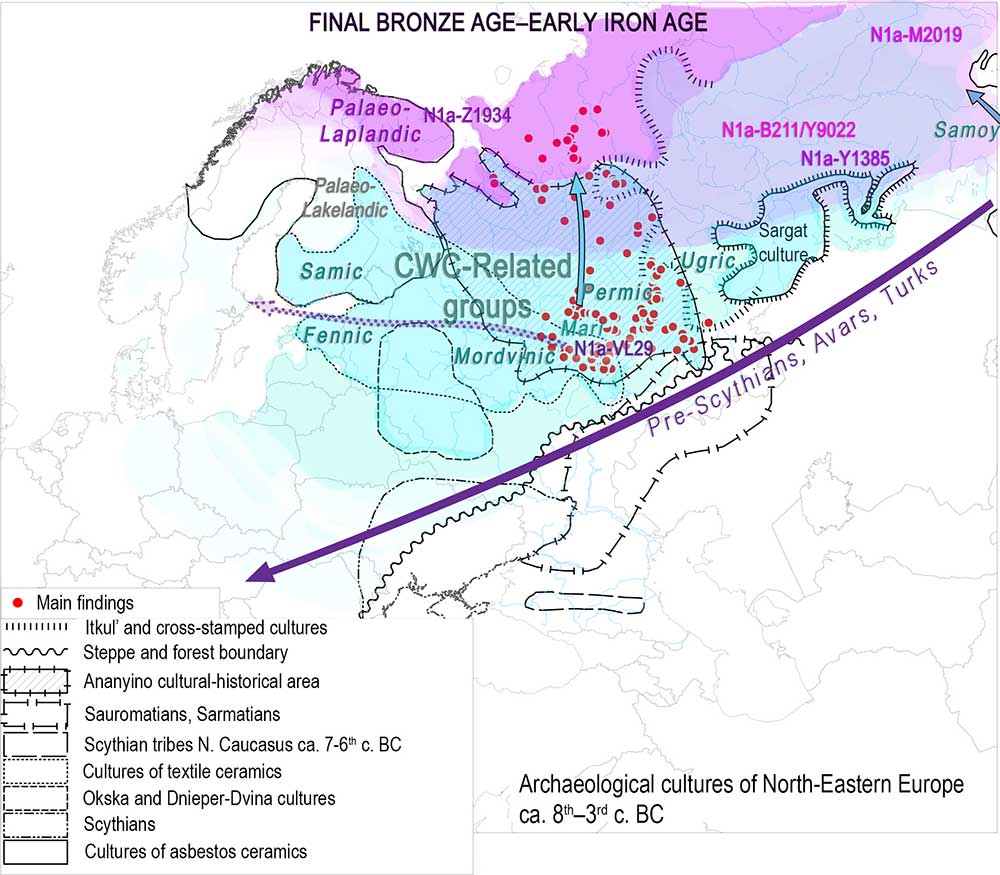
How did the ancient distribution or expansion of Palaeo-Arctic, Baikalic, and/or Altaic peoples affect the current distribution of the so-called “Siberian” ancestry, and of hg. N1a, in each specific population?
NOTE. A little excursus is necessary, because the calculated repetition of a hypothetic opposition “N1a vs. R1a” doesn’t make this dichotomy real:
- There was not a single ethnolinguistic community represented by hg. R1a after the initial expansion of Eastern Corded Ware groups, or by hg. N1a-L392 after its initial expansion in Siberia:
- there were R1a-Z645 subclades that became incorporated quite early into expanding Indo-Europeans (such as the R1a-Z93-rich Indo-Iranians), while others remained in Finno-Ugric communities until well into the Middle Ages;
- similarly, N1a-L392 subclades show a distinct ancient Palaeo-Siberian distribution including Altaic peoples to the south, and Arctic peoples to the north (more on this below).
- Different subclades became incorporated in different ways into Bronze Age and Iron Age communities, most of which without an ethnolinguistic change. For example, N1a subclades became incorporated into North Eurasian populations of different languages, reaching Uralic- and Indo-European-speaking territories of north-eastern Europe during the late Iron Age, at a time when their ancestral origin or language in Siberia was impossible to ascertain. Just like the mix found among Proto-Germanic peoples (R1b, R1a, and I1)* or among Slavic peoples (I2a, E1b, R1a)*, the mix of many Uralic groups showing specific percentages of R1a, N1a, or Q subclades* reflect more or less recent admixture or acculturation events with little impact on their languages.
*other typically northern and eastern European haplogroups are also represented in early Germanic (N1a, I2, E1b, J, G2), Slavic (I1, G2, J) and Finno-Permic (I1, R1b, J) peoples.
The problem with mapping the ancestry of the available sampling of ancient populations is that we lack proper temporal and regional transects. The maps that follow include cultures roughly divided into either “Bronze Age” or “Iron Age” groups, although the difference between samples may span up to 2,000 years.
NOTE. Rough estimates for more external groups (viz. Sweden Battle Axe/Gotland_A for the NW, Srubna from the North Pontic area for the SW, Arctic/Nganasan for the NE, and Baikal EBA/”Ulchi-like” for the SE) have been included to offer a wider interpolated area using data already known.
Bronze Age
Similar to modern populations, the selection of best fit “Siberian” ancestry between Baikal HG vs. Nganasan, both potentially ± ANE (AG3), is an oversimplification that needs to be addressed in future papers.
Corded Ware ancestry
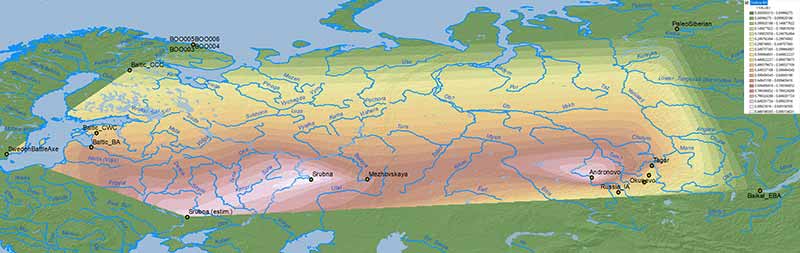
Nganasan-like ancestry
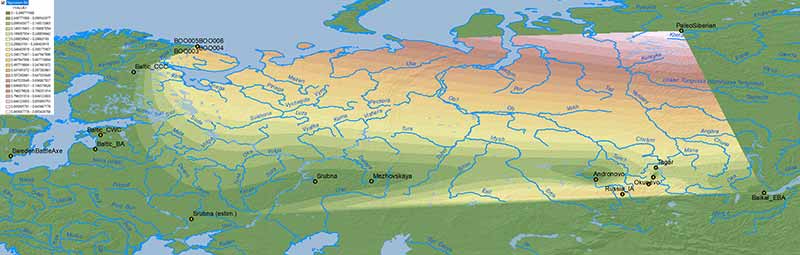
Baikal HG ancestry
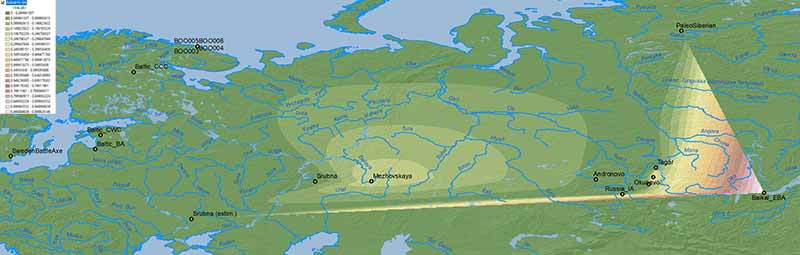
Afontova Gora 3 ancestry
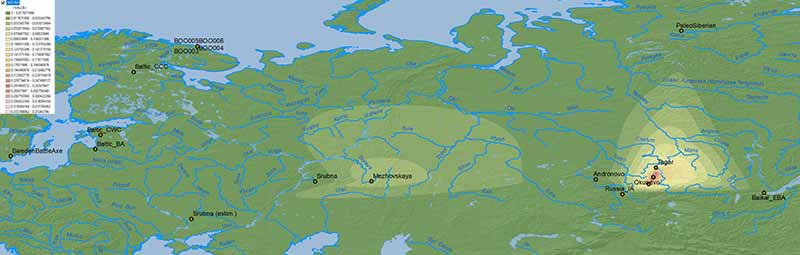
Iron Age
Corded Ware ancestry
Interestingly, the moderate expansion of Corded Ware-related ancestry from the south during the Iron Age may be related to the expansion of hg. N1a-VL29 into the chiefdom-based system of north-eastern Europe, including Ananyino/Akozino and later expanding Akozino warrior-traders around the Baltic Sea.
NOTE. The samples from Levänluhta are centuries older than those from Estonia (and Ingria), and those from Chalmny Varre are modern ones, so this region has to be read as a south-west to north-east distribution from the Iron Age to modern times.
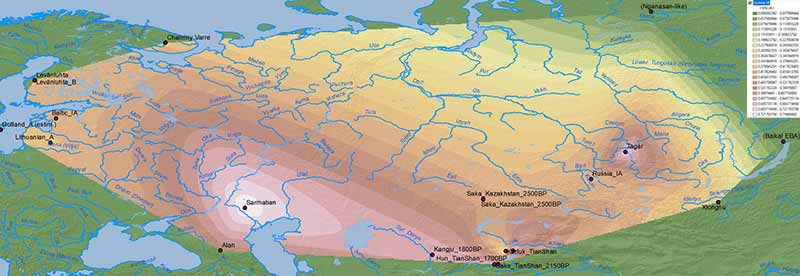
Baikal HG-like ancestry
The fact that this Baltic N1a-VL29 branch belongs in a group together with typically Avar N1a-B197 supports the Altaic origin of the parent group, which is possibly related to the expansion of Baikalic ancestry and Iron Age nomads:
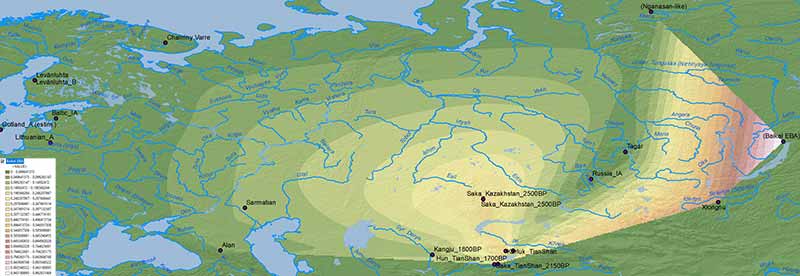
Nganasan-like ancestry
The dilution of Nganasan-like ancestry in an Arctic region featuring “Siberian” ancestry and hg. N1a-L392 at least since the Bronze Age supports the integration of hg. N1a-Z1934, sister clade of Ugric N1a-Z1936, into populations west and east of the Urals with the expansion of Uralic languages to the north into the Tundra region (see here).
The integration of N1a-Z1934 lineages into Finnic-speaking peoples after their migration to the north and east, and the displacement or acculturation of Saami from their ancestral homeland, coinciding with known genetic bottlenecks among Finns, is yet another proof of this evolution:
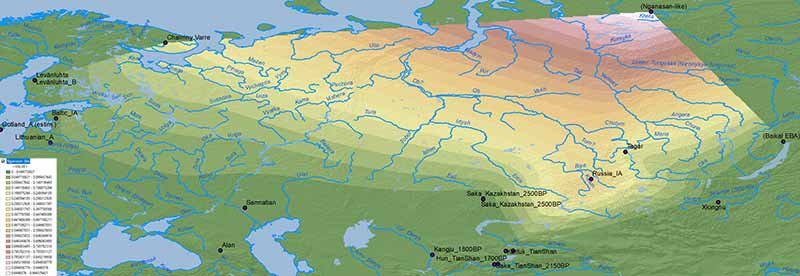
WHG ancestry
Similarly, WHG ancestry doesn’t seem to be related to important population movements throughout the Bronze Age, which excludes the multiple North Eurasian populations that will be found along the clines formed by WHG, EHG, ANE, Nganasan, Baikal HG ancestry as forming part of the Uralic ethnogenesis, although they may be relevant to follow later regional movements of specific populations.
Conclusion
It seems natural that people used to look at maps of haplogroup distribution from the 2000s, coupled with modern language distributions, and would try to interpret them in a certain way, reaching thus the wrong conclusions whose consequences are especially visible today when ancient DNA keeps contradicting them.
In hindsight, though, assuming that Balto-Slavs expanded with Corded Ware and hg. R1a, or that Uralians expanded with “Siberian” ancestry and hg. N1a, was as absurd as looking at maps of ancestry and haplogroup distribution of ancient and modern Native Americans, trying to divide them into “Germanic” or “Iberian”…
The evolution of each specific region and cultural group of North Eurasia is far from being clear. However, the general trend speaks clearly in favour of an ancient, Bronze Age distribution of North Eurasian ancestry and haplogroups that have decreased, diluted, or become incorporated into expanding Uralians of Corded Ware ancestry, occasionally spreading with inter-regional expansions of local groups.
Given the relatively recent push of Altaic and Indo-European languages into ancestral Uralic-speaking territories, only the ancient Corded Ware expansion remains compatible with the spread of Uralic languages into their historical distribution.
Related
- Uralic speakers formed clines of Corded Ware ancestry with WHG:ANE populations
- Vikings, Vikings, Vikings! “eastern” ancestry in the whole Baltic Iron Age
- European hydrotoponymy (IV): tug of war between Balto-Slavic and West Uralic
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Genetic continuity among Uralic-speaking cultures in north-eastern Europe
- Volosovo hunter-gatherers started to disappear earlier than previously believed
- Common Slavs from the Lower Danube, expanding with haplogroup E1b-V13?
- “Dinaric I2a” and the expansion of Common Slavs from East-Central Europe
- The cradle of Russians, an obvious Finno-Volgaic genetic hotspot
- Aquitanians and Iberians of haplogroup R1b are exactly like Indo-Iranians and Balto-Slavs of haplogroup R1a
- Consequences of O&M 2018 (III): The Balto-Slavic conundrum in Linguistics, Archaeology, and Genetics
- Pre-Germanic and Pre-Balto-Finnic shared vocabulary from Pitted Ware seal hunters
- Kortlandt: West Indo-Europeans along the Danube, Germanic and Balto-Slavic share a Corded Ware substrate
- Pre-Germanic born out of a Proto-Finnic substrate in Scandinavia
- On Proto-Finnic language guesstimates, and its western homeland
- On the origin and spread of haplogroup R1a-Z645 from eastern Europe
- Correlation does not mean causation: the damage of the ‘Yamnaya ancestral component’, and the ‘Future American’ hypothesis