Last modified: 1st November 2017
AdmixTools
Install admixtools
You can do it with git (you have to have it installed):
sudo apt-get install git
git clone https://github.com/DReichLab/AdmixTools.git
(you can add at the end the folder where you want it downloaded).
There you have the AdmixTools folder.
Another option, if you don’t like git, is wget (or just download the zip directly):
wget https://github.com/DReichLab/AdmixTools/archive/master.zip
unzip master.zip
And there you have the AdmixTools-master folder.
It has to be installed from source, and I had the following packages installed – however, probably just the basic packages (GSL, lapack and openblas, in bold) would have been enough, as stated in the software instructions (I would install at least those in bold):
sudo apt-get install gsl-bin libgsl-dbg libgsl-dev
(you might need to install libgsl0-dbg and libgsl0-dev instead)
sudo apt-get install gfortran liblapack3 liblapack-dev liblapack-doc liblapack-pic liblapacke liblapacke-dev
sudo apt-get install libblas3 libblas-common libblas-dev libblas-doc libatlas3-base libatlas-base-dev libblas-test libopenblas-base libopenblas-dev
To install (per the instructions in the README file), from the /src/ folder:
cd AdmixTools/src
make clobber
make install
If you encounter some strange problems, follow the instructions of the README:
wget https://reich.hms.harvard.edu/sites/reich.hms.harvard.edu/files/inline-files/AdmixTools_Example_Data.tar_.gz
tar -zxvf AdmixTools_Example_Data.tar_.gz
The extracted folder /data/ should be in the AdmixTools/ or AdmixTools-master/ folder. Now from the /examples/ folder run the examples:
cd AdmixTools/examples
./mklog
If you encounter problems, it is probably related to your system.
Using ADMIXTOOLS
The documentation is pretty straightforward – at least if you are able to use other instructions, like those for PCA and ADMIXTURE.
You can see examples from this blog and useful instructions from other websites:
- GAWorkshop Outgroup F3 Statistics detailed instructions.
- ADMIXTOOLS documentation – read the specific README.XXX files for the software tool you want to use
Samples
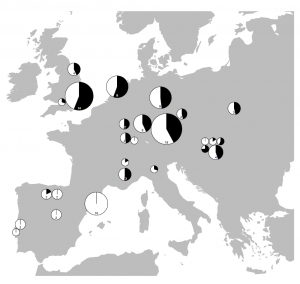
Survival of hunter-gatherer ancestry in West-Central European Neolithic
R1b-L23-rich Bell Beaker-derived Italic peoples from the West vs. Etruscans from the East
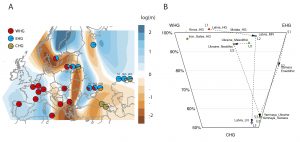
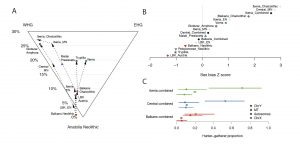
“Steppe ancestry” step by step (2019): Mesolithic to Early Bronze Age Eurasia
Bell Beakers and Mycenaeans from Yamnaya; Corded Ware from the forest steppe
Samples used in the Indo-European demic diffusion model:
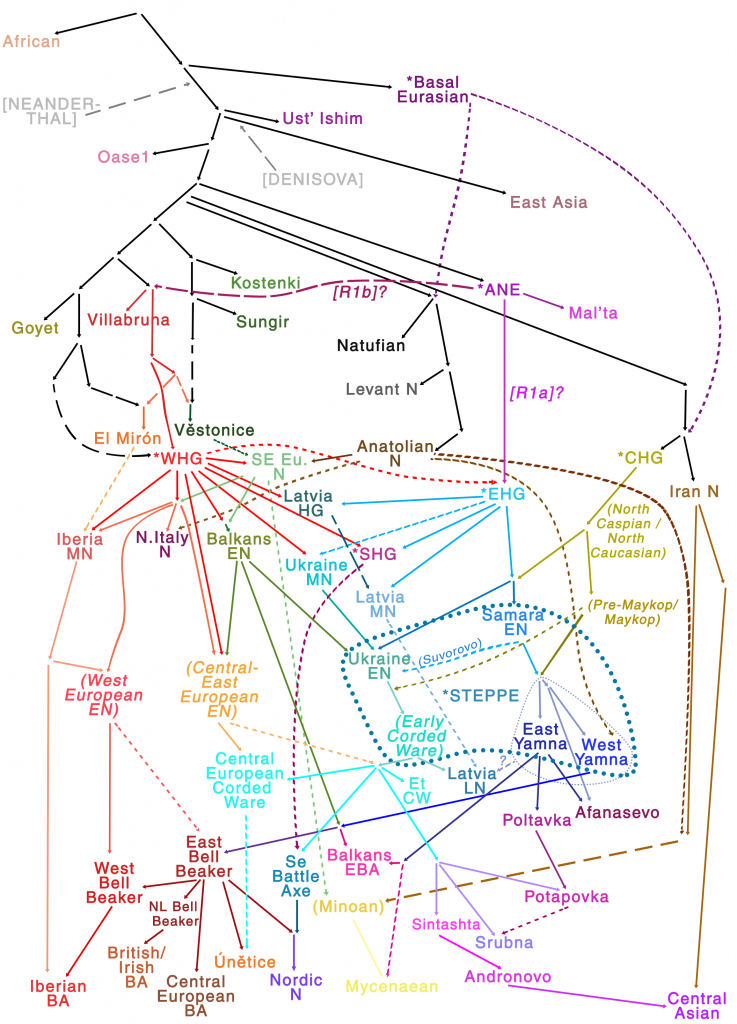
Other samples included in the model, from recent genetic papers: