Since 2018, I have compiled data of Y-DNA and mtDNA for most reported ancient samples, including analyses of BAM files by hobbyists and online informal reports of research papers in preparation, including also automated haplogroup inferences performed with the software Yleaf v.2 used by professional geneticists. I referenced the first version of the dataset in the book series A Song of Sheep and Horses, and I have improved it a lot since then, in quantity and in quality.
You can access the latest version of the data at the following links:
![]() Google Drive folder with the latest version of the files.
Google Drive folder with the latest version of the files.
![]() As shortcut, just type haplogroup.info on your browser.
As shortcut, just type haplogroup.info on your browser.
NOTE. You can download the files or read them online. Especially for slower computers, it is recommended to read and search online the native Google Spreadsheet. The behaviour of the original properties of the Excel sheet might differ when reading it with Google Drive, though.
Direct link to files from haplogroup.info (may not be the latest ones):
 Excel: original file.
Excel: original file. Excel: lightweight version.
Excel: lightweight version. CSV: comma-separated file (non-UTF8 to avoid errors).
CSV: comma-separated file (non-UTF8 to avoid errors). TSV: tab-separated values (text file).
TSV: tab-separated values (text file). PDF for reading and searching.
PDF for reading and searching. HTML for online reference (slow to load!).
HTML for online reference (slow to load!).
For more information on the meaning of columns, color codes, emphasis, and styles in general check the second sheet in the file, called supp-info. For a full reference of the papers and accession numbers, check the third sheet called papers.
By default, samples are ordered by ISOGG nomenclature and (secondarily) by mean year cal. BC, which gives a more natural visual of Y-DNA subclades, but you can order samples by any other column, search for specific values, etc.
A formatted spreadsheet with all FTDNA SNPs available to date has been kindly provided by Göran Runfeldt.
Updates with their date and comment start anew in major versions. For updates, comments, etc. there is a dedicated thread at the forum.
Announcements and links to new Y-DNA SNP calls will be made available from the appropriate section of the forum.
Citing this work
You can reuse and modify the files posted here and their content as you see fit, for any personal or academic research, as well as for any kind of project, whether personal or professional, open or copyrighted.
 Copyright 2018-2020 Carlos Quiles.
Copyright 2018-2020 Carlos Quiles. Copyright 2013-2018 Jean Manco.
This work is licensed under a Creative Commons Attribution 3.0 Unported License.
Please always cite the source with version number, and whenever possible (online work) this website indo-european.eu, too.
All samples including haplogroup inference different from the original publication have an individual “responsible”. If you find that the inference or its author is incorrect, please contact me.
NOTE. I have checked some of these inferences – many more than the most relevant ones I posted here – and, whenever necessary, selected those I agree with. My own inferences follow results from YLeaf v.2
Beyond the simplest netiquette, you should cite and/or link to this file whenever you publish any bulk information remade or taken from (or based on) it, for a simple reason:
If you reference the file with its version, you make it easier for people to track changes, especially those related to errors corrected since you last downloaded or checked it, and you offer thus information on how reliable your data (and project) actually is.
Collaboration
Everyone can read the online spreadsheet, but only I am able to modify it. The best way to make changes is to email me corrections directly at [email protected].
Alternatively, you can leave a comment on this page or any post of this blog related to specific samples, or at the Indo-European forum, especially if you think it merits some discussion.
If you have an interesting (public or private) project and want to receive immediate notifications of updates from Google Drive, or you would like to discuss the possibility of editing the file directly for bulk modifications, you may also contact me.
Maps and GIS
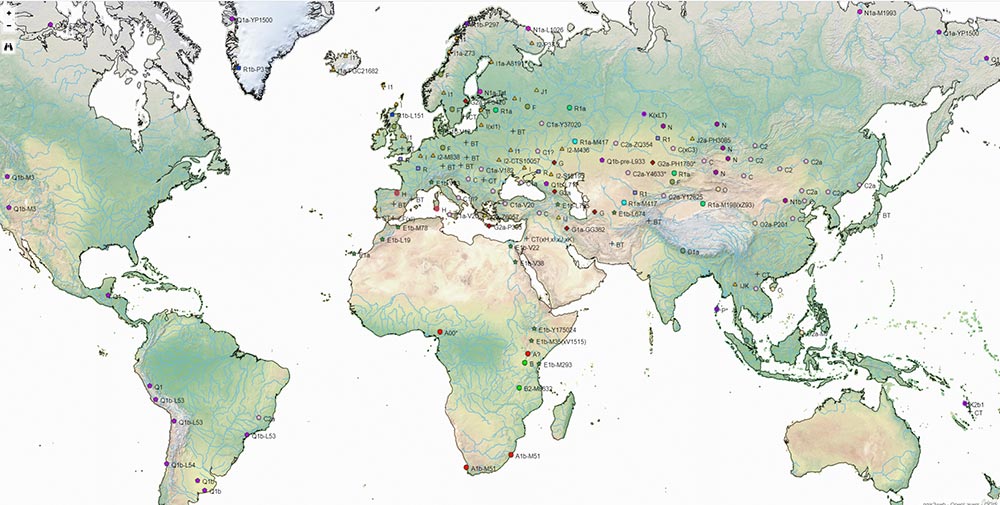
ArcGIS Online
ArcGIS Online offers the possibility of publishing data layers over image layers with predetermined themes for ease of use. You can read more about instructions for use of the software.
NOTE. The easier to remember domain name haplogroup.info can be used to access both, this ArcGIS Online map and the Google Drive folder with the dataset.
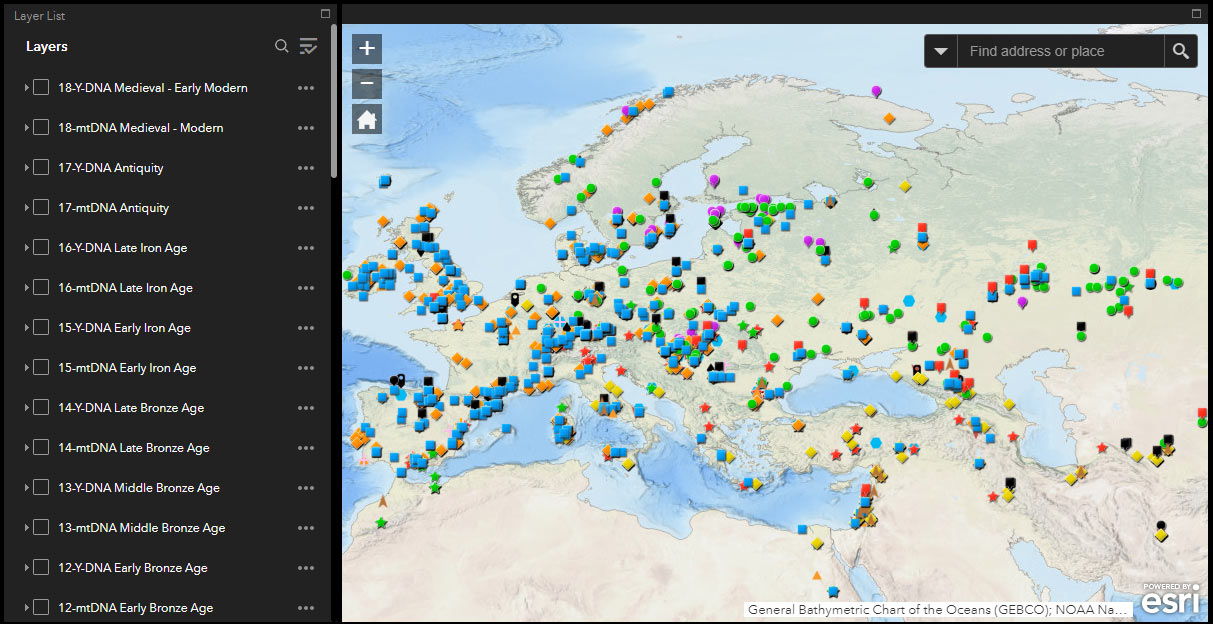
QGIS
The free GIS software QGIS offers the possibility to export different open formats, so I have used it to publish online maps with all samples and also divided by age, that users can quickly refer to.
There is an updated list of available static and dynamic maps for reference of Y-DNA and mtDNA haplogroups of ancient samples:
Other Projects
I will post here relevant projects using this spreadsheet:
NOTE. Before you do a simple copy (or just some minor modifications) to publish it elsewhere, think about how your contribution is going to compensate for the multiple errors that will be corrected in this spreadsheet, while your copy (and many other similar ones) might keep spreading them. In my experience, it becomes very soon prohibitive to track all changes done to just two files, so imagine multiple ones. If your aim is to improve certain parts, think about collaborating instead.
SNP Tracker
SNP Tracker is currently the only free tool to obtain migration paths of SNPs, offering inferences based on FTDNA SNPs and additional YFull data at a professional level.
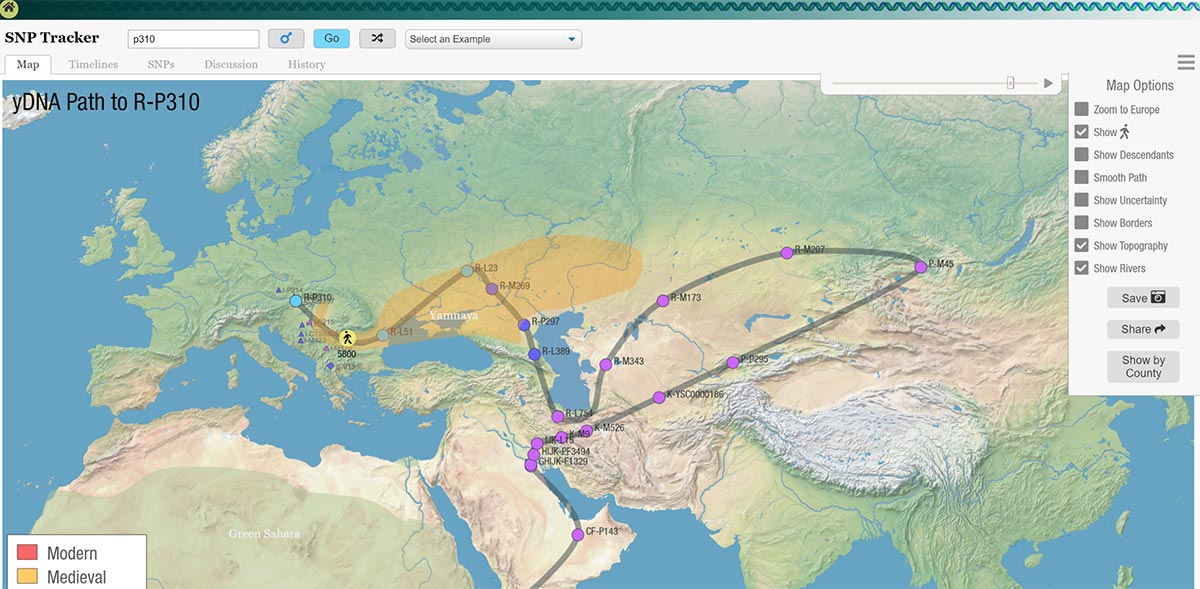
His author, Rob Spencer, is a retired MIT graduate (see his CV), and is in continuous contact with other knowledgeable researchers linked to FTDNA projects.
All maps are created on the fly in JavaScript in the user’s browser, and graphics incorporate both raster and vector together according to the user’s preferences.
The tool is constantly updated and uses YFull SNP formation dates to interpolate dates onto the much larger FTDNA Y and mt SNP trees. SNP Tracker loads the full Y and mt trees in the background.
Furthermore, Paleolithic to Bronze Age paths are largely determined by hand-curated SNPs that are “pegged” to specific locations, avoiding thus the usual distortions of any algorithm blindly applied to the data. To validate these “pegged” locations, Rob has analysed DNA data from ancient sites here relative to specific SNPs to see how they work out compared to other automatic means.
NOTE. For example, it is very likely that, without “pegging”, R1b’s route based on modern samples would probably follow a maritime route from the Middle East through the Mediterranean into Central Europe…
Maps also show visually additional information, such as plagues, cultures, other “co-resident” haplogroups, etc. without excess visual clutter.
Rob is now actively developing new ideas based on ancient DNA data, so stay tuned to his website for news!
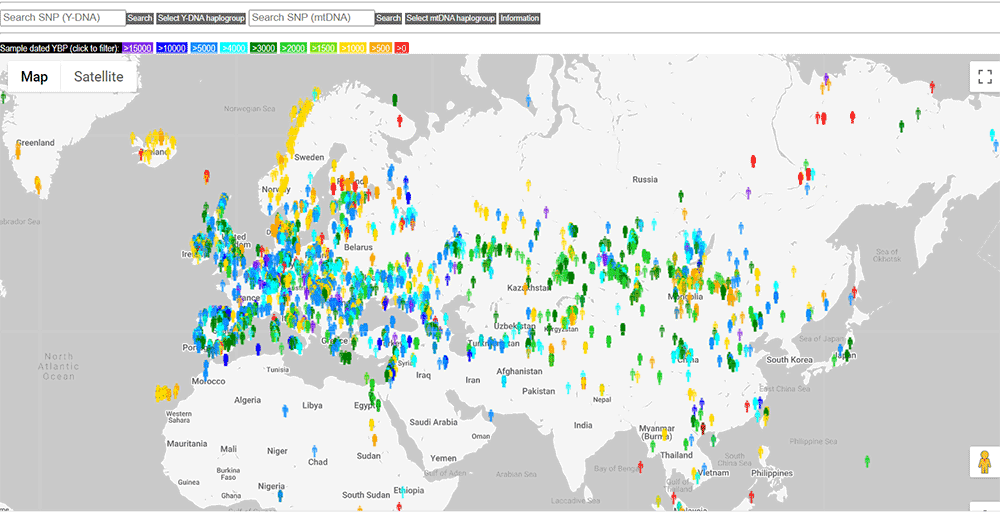
HaploTree
As part of the Haplotree Information Project, the AncientDNA.info project incorporates ancient DNA for easy search and reference of Y-DNA and mtDNA haplogroups, including a color code for quick visualization of their chronology.
new sample from levant and other study That you did not mention)Responsible-SNP:
Please Update Database
https://anthrogenica.com/showthread.php?20521-Y-Chromosome-Haplogroup-assignments-for-2500-ancient-samples
https://drive.google.com/file/d/1vAyEHt7x4ufYxAzn8audhyBe1vgNUI_n/view
Thank you very much, this is cool. I’ve added most subclades not already available on the Dataset and updated the maps.
The file is great because it offers positive SNPs potentially not reported by the papers. Even those from the recent paper on SE Asia, which didn’t include Y-DNA, or others from which we only have genotype.
The problems are:
On the outside, comparing it with previous assignments, it does a fairly good job, so I could trust it to compare and automatically update haplogroups.
But, it will need a lot of attention by interested users looking at the reported SNPs in the specific files and possibly analyzing the BAMs directly to catch potential errors and improve subclade assignment. I can’t analyze in detail 2500 samples…
Hello
Dear carlos
Please update
Sample I2335 (Low Coverage) is probably related to J2
The accompanying Samples are also J2
https://www.mediafire.com/file/dwe9ihh6gljs7so/I2335.xlsx
Thank You
Hola Carlos: ArcGIS Online Map de https://haplogroup.info/ no funciona hace días que no funciona.
Un cordial saludo,
Ariel
Hi!
Thank you for your great effort!
I have a small problem with the map. Every time I want to open the link of the map, it reminds me to login. But every time I enter the name and password, it goes wrong. I am sure my account is fine because I can comment now. Could you please show me how to solve the problem?
Thank you!
Hi
Please add new sarmatian
https://www.sciencedirect.com/science/article/pii/S0092867422002677
and
https://www.academia.edu/15713987