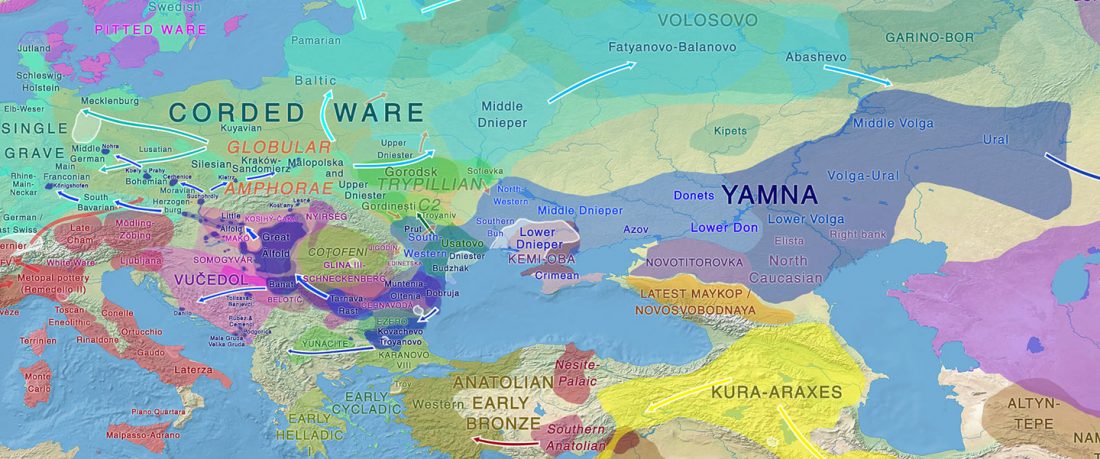
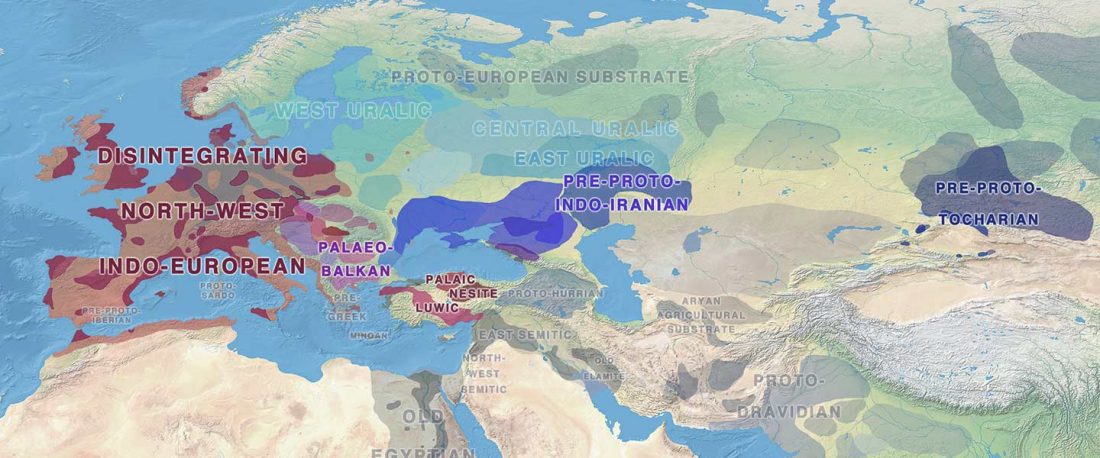
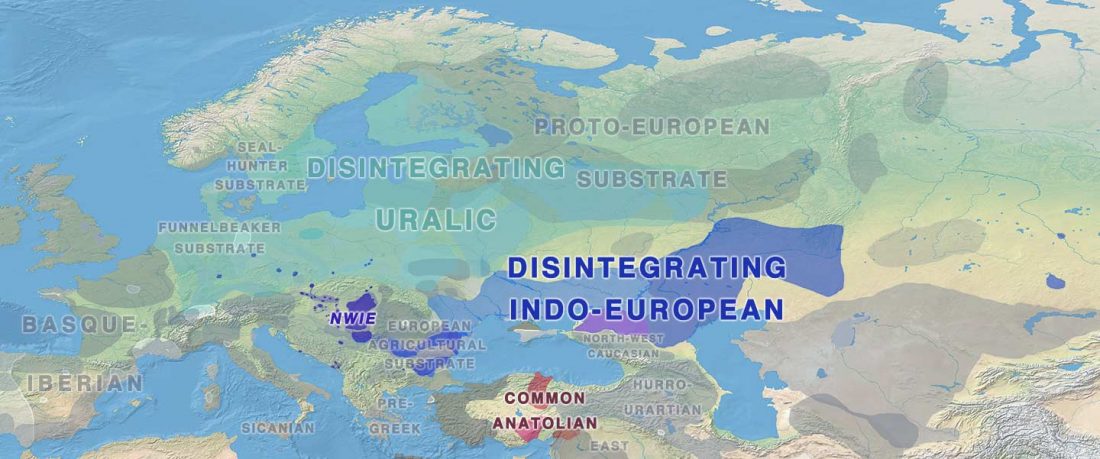
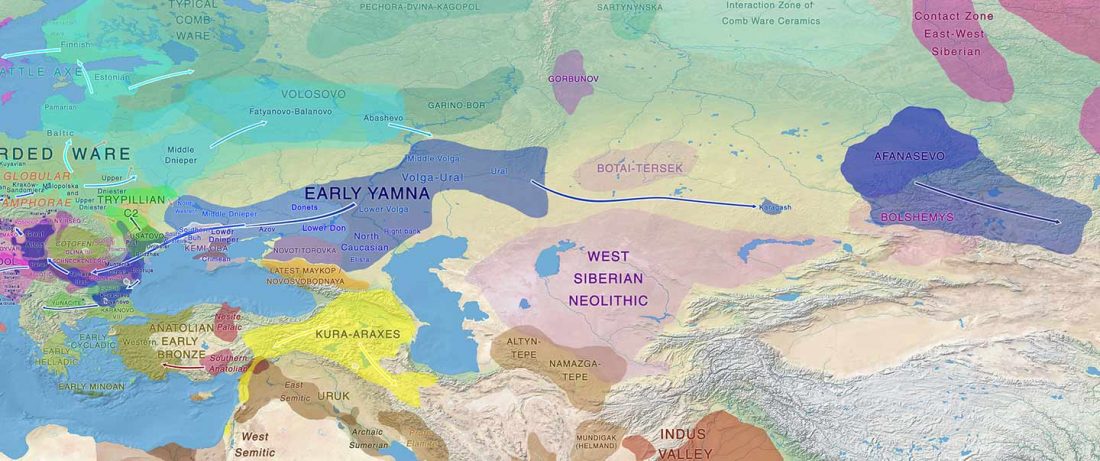
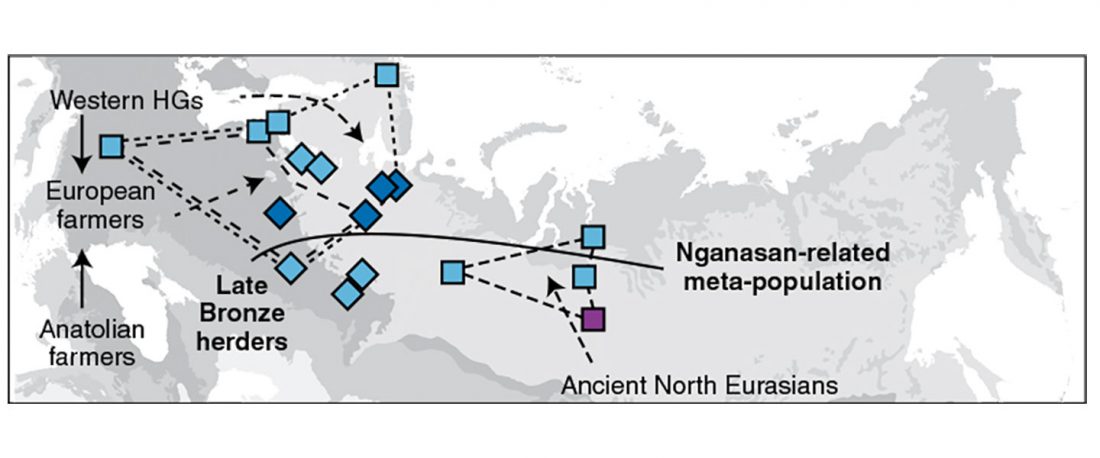
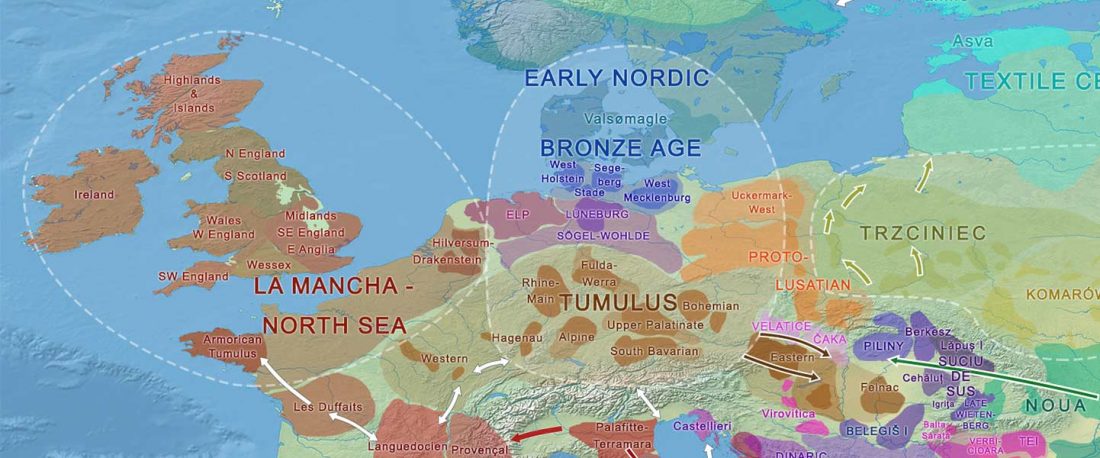
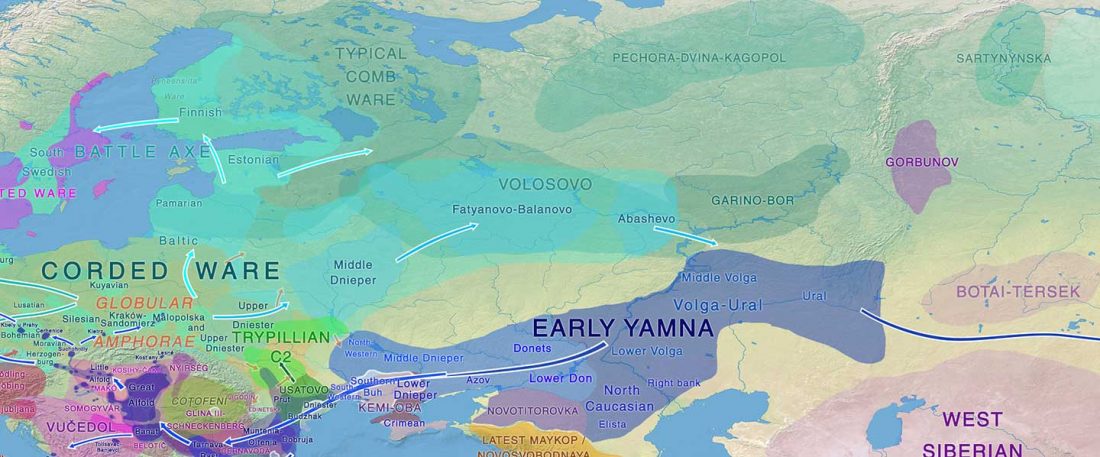
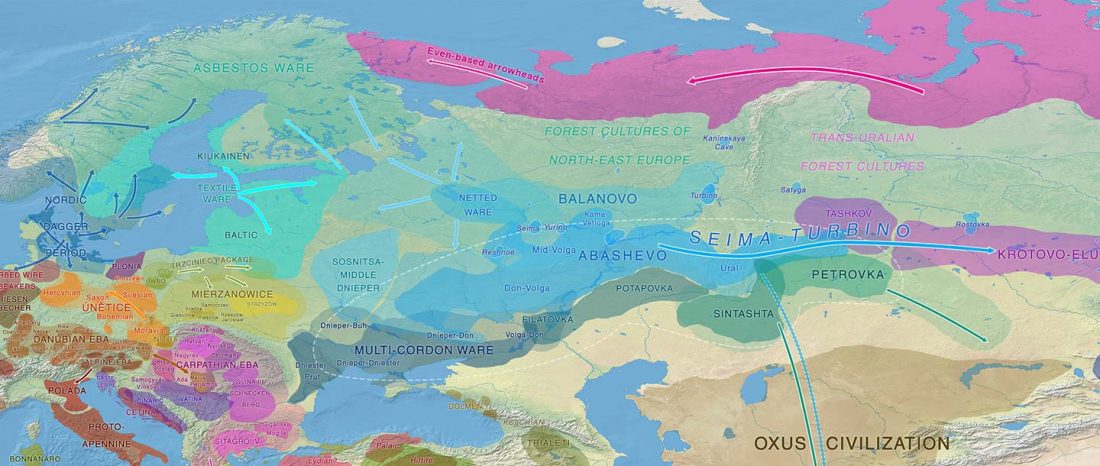
Recently, the preprint by Sirak et al. biorXiv (2019), Human auditory ossicles as an alternative optimal source of ancient DNA, was published in Genome Res. (2020), and the corresponding samples were finally uploaded to ENA.
I have been trying to get my hands on sample GLAV_14, a male from the Late Eneolithic site Glăvăneştii Vechi, classified as Romania Bronze Age (ca. 3500-3000 BC), mtDNA T1a1, referenced as investigated first in the study:
… Read the rest “Earliest R1a-Z93…from Late Trypillia in the Podolian-Volhynian Upland!”Haas N, Maximilian K. 1958. Anthropological study of the human bones from graves with ochre from Glăvăneștii Vechi, Corlăteni and Stoicani Cetățuie. Soviet Anthropology 4,