Two important papers have appeared regarding the supposed link of Uralians with haplogroup N.
Avars of haplogroup N1c-Tat
Preprint Genetic insights into the social organisation of the Avar period elite in the 7th century AD Carpathian Basin, by Csáky et al. bioRxiv (2019).
Interesting excerpts (emphasis mine):
After 568 AD the Avars settled in the Carpathian Basin and founded the Avar Qaganate that was an important power in Central Europe until the 9th century. Part of the Avar society was probably of Asian origin, however the localisation of their homeland is hampered by the scarcity of historical and archaeological data.
Here, we study mitogenome and Y chromosomal STR variability of twenty-six individuals, a number of them representing a well-characterised elite group buried at the centre of the Carpathian Basin more than a century after the Avar conquest.
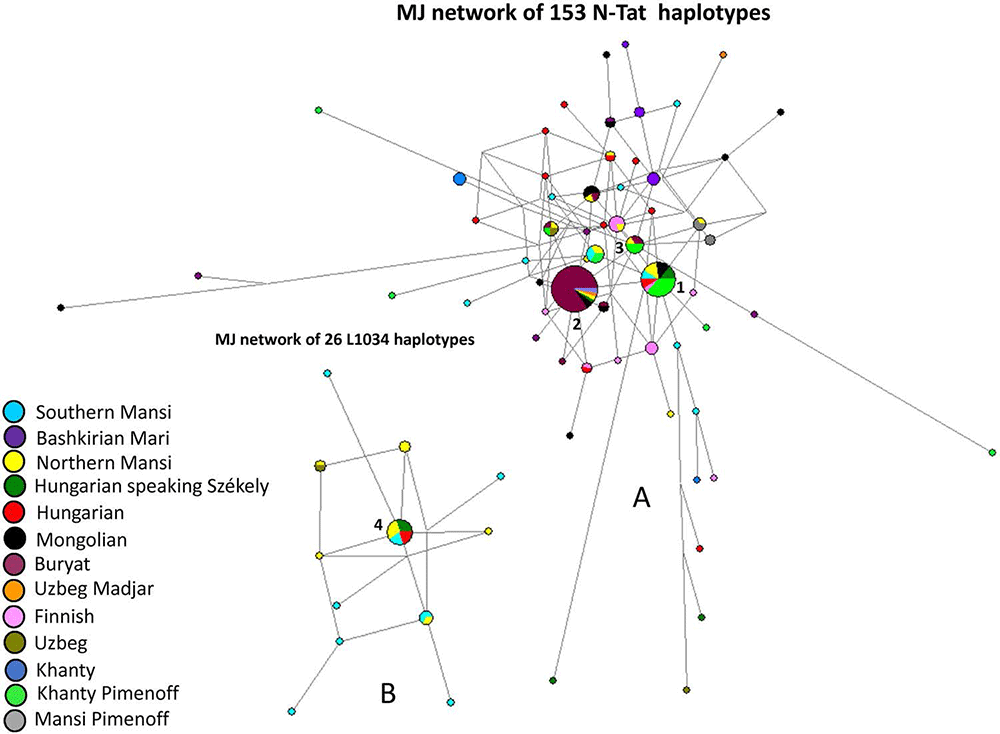
The Y-STR analyses of 17 males give evidence on a surprisingly homogeneous Y chromosomal composition. Y chromosomal STR profiles of 14 males could be assigned to haplogroup N-Tat (also N1a1-M46). N-Tat haplotype I was found in four males from Kunpeszér with identical alleles on at least nine loci. The full Y-STR haplotype I, reconstructed from AC17 with 17 detected STRs, is rare in our days. Only nine matches were found among haplotypes in YHRD database, such as samples from the Ural Region, Northern Europe (Estonia, Finland), and Western Alaska (Yupiks). We performed Median Joining (MJ) network analysis using N-Tat haplotypes with ten shared STR loci (Fig. 3, Table S9). All modern N-Tat samples included in the network had derived allele of L708 as well. Haplotype I (Cluster 1 in Fig. 3) is shared by eight populations on the MJ network among the 24 identical haplotypes. Cluster 1 represents the founding lineage, as it is described in Siberian populations, because this haplotype is shared by the most populations and it is more diverse than Cluster 2.
Nine males share N-Tat haplotype II (on a minimum of eight detected alleles), all of them buried in the Danube-Tisza Interfluve. We found 30 direct matches of this N-Tat haplotype II in the YHRD database, using the complete 17 STR Y-filer profile of AC1, AC12, AC14, AC15, AC19 samples. Most hits came from Mongolia (seven Buryats and one Khalkh) and from Russia (six Yakuts), but identical haplotypes also occur in China (five in Xinjiang and four in Inner Mongolia provinces). On the MJ network, this haplotype II is represented by Cluster 2 and is composed of 45 samples (including 32 Buryats) from six populations (Fig. 3).
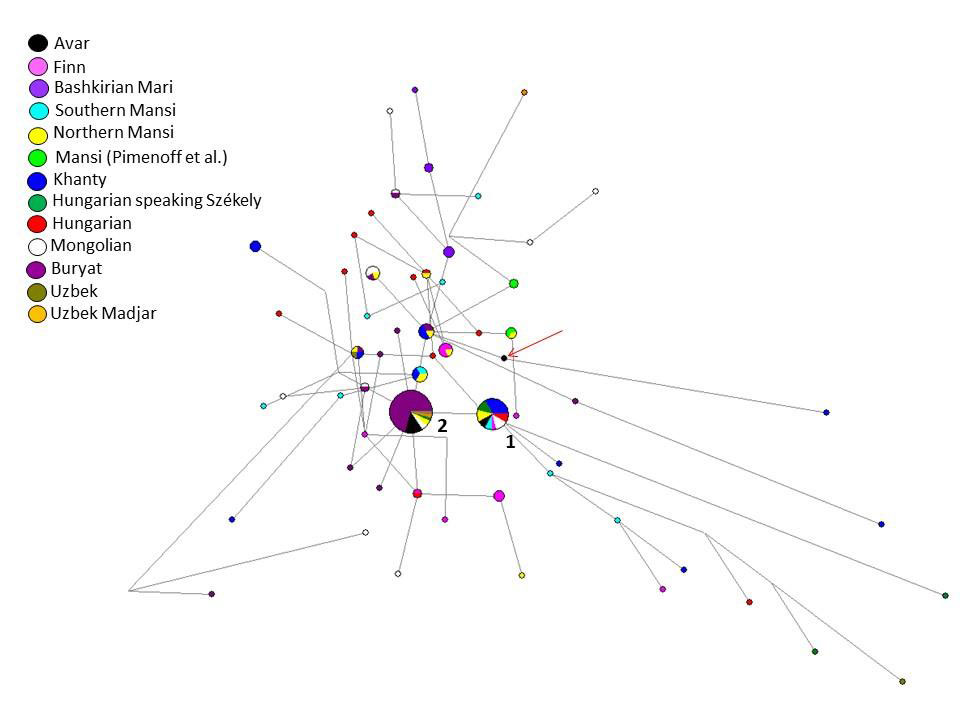
A third N-Tat lineage (type III) was represented only once in the Avar dataset (AC8), and has no direct modern parallels from the YHRD database. This haplotype on the MJ network (see red arrow in Fig. 3) seems to be a descendent from other haplotype cluster that is shared by three populations (two Buryat from Mongolia, three Khanty and one Northern Mansi samples). This haplotype cluster also differs one molecular step (locus DYS393) from haplotype II. We classified the Avar samples to downstream subgroup N-F4205 within the N-Tat haplogroup, based on the results of ours and Ilumäe et al.18 and constructed a second network (Fig. S4). The N-F4205 network results support the assumption that the N-Tat Avar samples belong to N-F4205 subgroup (see SI chapter 1d for more details).
Based on our calculation, the age of accumulated STR variance (TMRCA) within N-Tat lineage for all samples is 7.0 kya (95% CI: 4.9 – 9.2 kya), considering the core haplotype (Cluster 1) to be the founding lineage. Y haplogroup N-Tat was not detected by large scale Eurasian ancient DNA studies but it occurs in late Bronze Age Inner Mongolia and late medieval Yakuts, among them N-Tat has still the highest frequency.
Two males (AC4 and AC7) from the Transtisza group belong to two different haplotypes of Y-haplogroup Q1. Both Q1a-F1096 and Q1b-M346 haplotypes have neither direct nor one step neighbour matches in the worldwide YHRD database. A network of the Q1b-M346 haplotype shows that this male had a probable Altaian or South Siberian paternal genetic origin.
EDIT (5 APR 2019): The paper offers an interesting late sample before the arrival of Hungarian conquerors, although we don’t know which precise lineage the sample belongs to:
One sample in our dataset (HC9) comes from this population, and both his mtDNA (T1a1b) and Y chromosome (R1a) support Eastern European connections. (…) Furthermore, we excluded sample HC9 from population-genetic statistical analyses because it belongs to a later period (end of 7th – early 9th centuries)
Apparently, then, results are consistent with what was already known from studies of modern populations:
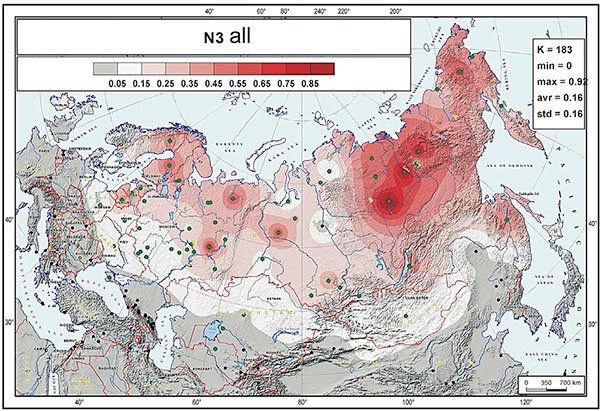
According to Ilumäe et al. study, the frequency peak of N-F4205 (N3a5-F4205) chromosomes is close to the Transbaikal region of Southern Siberia and Mongolia, and we conclude that most Avar N-Tat chromosomes probably originated from a common source population of people living in this area, completely in line with the results of Ilumäe et al.
Finno-Ugrians share haplogroup R1a-Z280
Another paper, behind paywall, Genetic history of Bashkirian Mari and Southern Mansi ethnic groups in the Ural region, by Dudás et al. Molecular Genetics and Genomics (2019).
Interesting excerpts (emphasis mine):
Y‑chromosome diversity
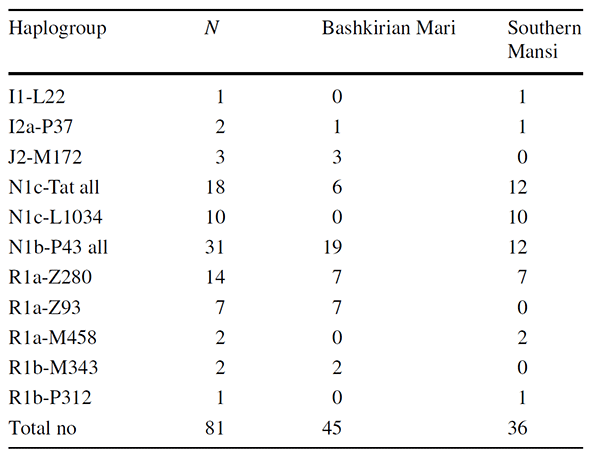
The most frequent haplogroups of the Bashkirian Maris were N1b-P43 (42%), R1a-Z280 (16%), R1a-Z93 (16%), N1c-Tat (13%), and J2-M172 (7%). Furthermore, subgroup R1b-M343 accounted for 4% and I2a-P37 covered 2% of the lineages. None of the Mari N1c Y chromosomes belonged to the N1c subgroups investigated (L1034, VL29, Z1936).
In the case of the Southern Mansi males, the most frequent haplogroups were N1b-P43 (33%), N1c-L1034 (28%) and R1a-Z280 (19%). The frequencies of the remaining haplogroups were as follows: R1a-M458 (6%), I1-L22 (3%), I2a-P37 (3%), and R1b-P312 (3%). The haplotype and haplogroup diversities of the Bashkirian Mari group were 0.9929 and 0.7657, whereas these values for the Southern Mansi were 0.9984 and 0.7873, respectively. The results show that, in both populations, haplotypes are much more diverse than haplogroups.
Genetic structure
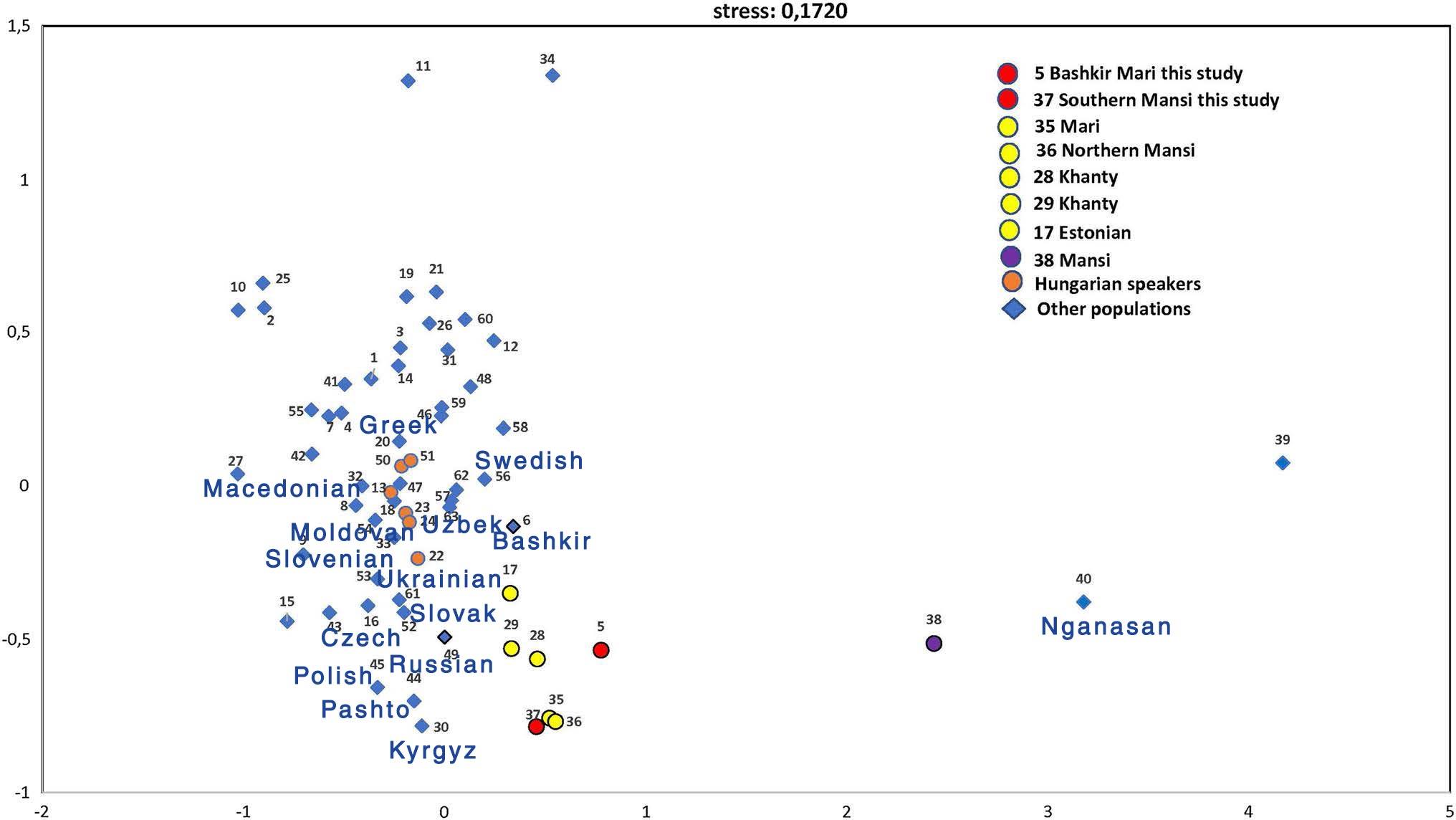
(..) the studied Bashkirian Mari and Southern Mansi population groups formed a compact cluster along with two Khanty, Northern Mansi, Mari, and Estonian populations based on close Fst-genetic distances (< 0.05), with nonsignificant p values (p > 0.05) except for the Estonian population. All of these populations belong to the Finno-Ugric language family. Interestingly, the other Mansi population studied by Pimenoff et al. (2008) (pop # 38) was located a great distance from the Southern Mansi group (0.268). In addition, the Bashkir population (pop # 6) did not show a close genetic affinity to the Bashkirian Mari group (0.194), even though it is the host population. However, the Russian population from the Eastern European region of Russia (pop # 49) showed a genetic distance of 0.055 with the Southern Mansi group. All Hungarian speaking populations (pops 13, 22, 23, 24, 50, and 51) showed close genetic affinities to each other and to the neighbouring populations, but not to the two studied populations.
Phylogenetic analysis
Median-joining networks were constructed for:
N-P43 (earlier N1b):
(…) TMRCA estimates for this haplogroup were made for all P43 samples (n = 157) 8.7 kya (95% CI 6.7–10.8 kya), for the N-P43 Asian.
N1c-Tat:
(…) 75% of Buryats belonged to Haplotype 2, indicating that the Buryats studied by us is a young and isolated population (Bíró et al. 2015). Bashkirian Mari samples derive from Haplotype 2 via Haplotype 3 (see dark purple circles on the top of Fig. 6a). Haplotype 3 contained six males (2 Buryat, 1 Northern Mansi, and 3 Khanty samples from Pimenoff et al. 2008). The biggest Bashkirian Mari haplotype node (3 Mari samples) was positioned three mutational steps away from Haplotype 1 and the remaining Mari samples can be derived from this haplotype. Southern Mansi haplotypes were scattered within the network except for two, which formed a smaller haplotype node with two Northern Mansi and two Khanty samples from Pimenoff et al. (2008).
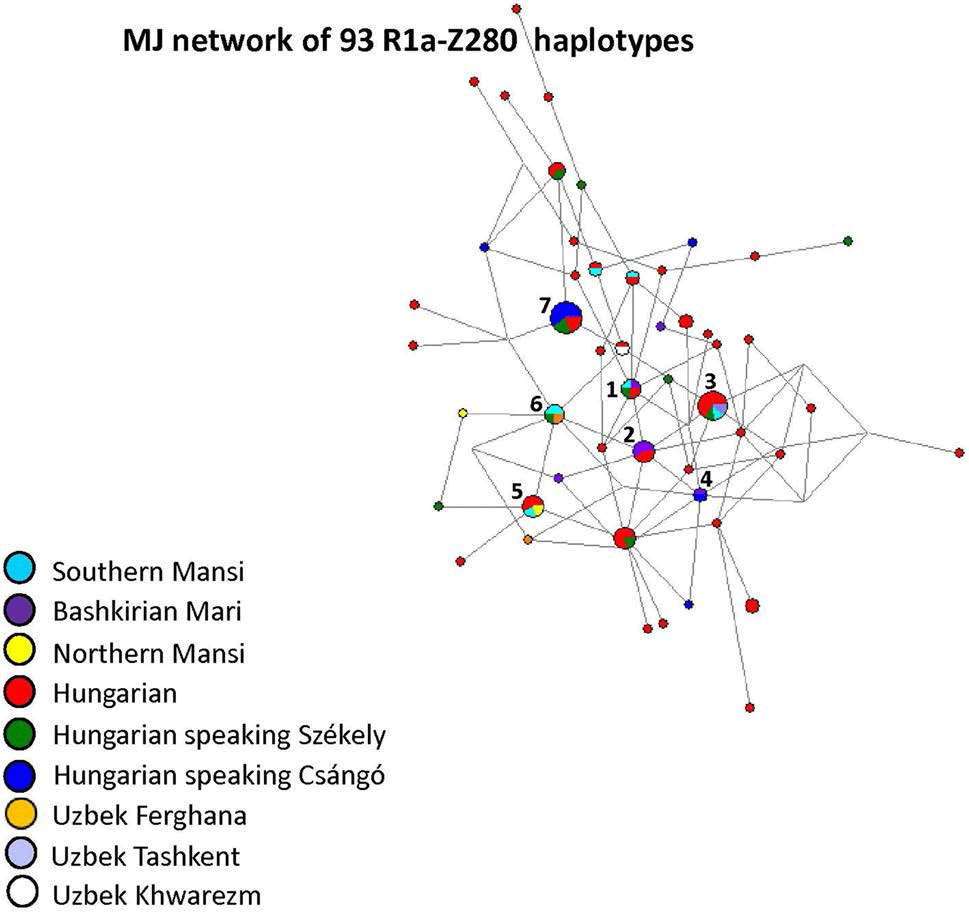
R1a-Z280 haplotypes, shared by Maris, Mansis, and Hungarians, hence ancient Finno-Ugrians:
The founder R1a-Z280 haplotype was shared by four samples from four populations (1 Bashkirian Mari; 1 Southern Mansi; 1 Hungarian speaking Székely; and 1 Hungarian), as presented in Fig. 7 (Haplotype 1). Haplotype 2 included five males (3 Bashkirian Mari and 2 Hungarian), as it can be seen in Fig. 7. Haplotype 4 included two shared haplotypes (1 Bashkirian Mari and one Hungarian speaking Csángó). The remaining two Bashkirian Mari haplotypes differ from the founder haplotype (Haplotype 1) by two mutational steps via Hungarian or Hungarian and Bashkirian Mari shared haplotypes. Beside Haplotype 1, the remaining Southern Mansi haplotypes were shared with Hungarians (Haplotype 5 or turquoise blue and red-coloured circles above Haplotype 7) or with Hungarians and Hungarian speaking Székely group (Haplotypes 3, 5, and 6). Haplotype 7 included ten Hungarian speakers (Hungarian, Székely, and Csángó). One Hungarian and one Uzbek Khwarezm shared haplotype can be found in Fig. 7 as well (red and white-coloured circle). All the other haplotypes were scattered in the network. The age of accumulated STR variation within R1a-Z280 lineage for 93 samples is estimated to be 9.4 kya (95% CI 6.5–12.4 kya) considering Haplotype 1 (Fig. 7) to be the founder.
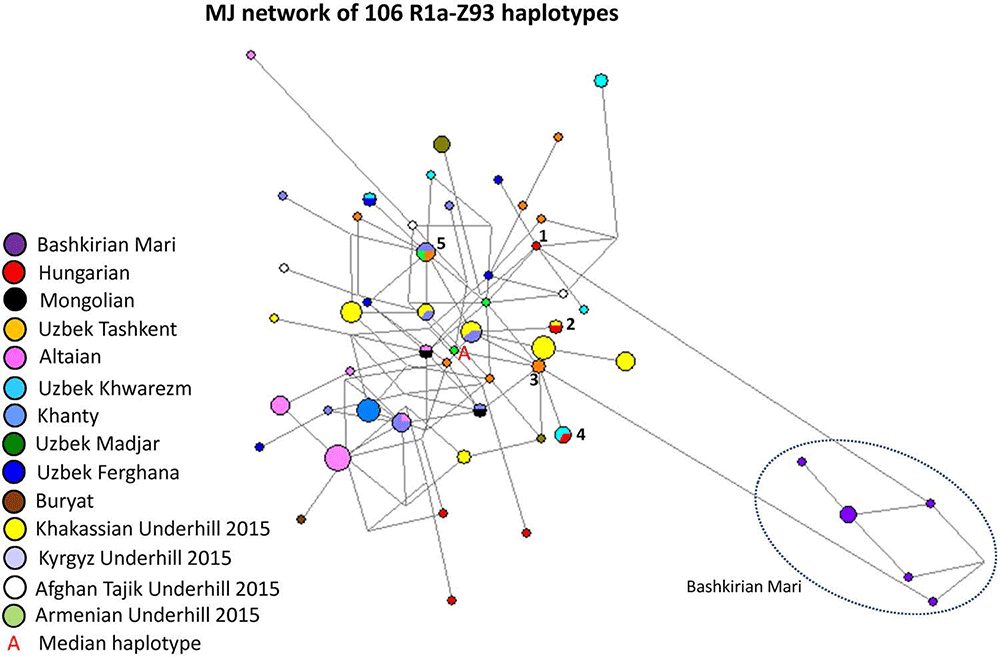
R1a-Z93 as isolated lineages among Permic and Ugric populations:
Figure 8 depicts an MJ network of R1a-Z93* samples using 106 haplotypes from the 14 populations (Fig. 8). All of the Bashkirian Mari samples (7 haplotypes) formed a very isolated branch and differed from the one Hungarian haplotype (Fig. 8, see Haplotype 1) by seven mutational steps as well from two Uzbek Tashkent samples (see Haplotype 3). Another Hungarian sample shared two haplotypes of Uzbek Khwarezm samples in Haplotype 4. This haplotype can be derived from Haplotype 3 (Uzbek Tashkent). Haplotype 2 included one Hungarian and one Khakassian male. The remaining three Hungarian haplotypes are outliers in the network and are not shared by any sample. The other population samples included in the network either form independent clusters such as Altaians, Khakassians, Khanties, and Uzbek Madjars or were scattered in the network. The age of accumulated STR variation (TMRCA) within R1a-Z93* lineage for 106 samples is estimated as 11.6 kya (95% CI 9.3–14.0 kya) considering an Armenian haplotype (Fig. 8, “A”) to be the founder and the median haplotype.
16 Altaian; 24 Khakassian; 12 Kyrgyz) from Underhill et al. (2015)
Comments
The results of modern populations for N (especially N1c) subclades show really wide clusters and ancient TMRCA, consistent with their known ancient and wide distribution in northern and eastern Eurasian groups, and thus with infiltration of different lineages with eastern nomads (and northern Arctic populations) coupled with later bottlenecks, as well as acculturation of groups.
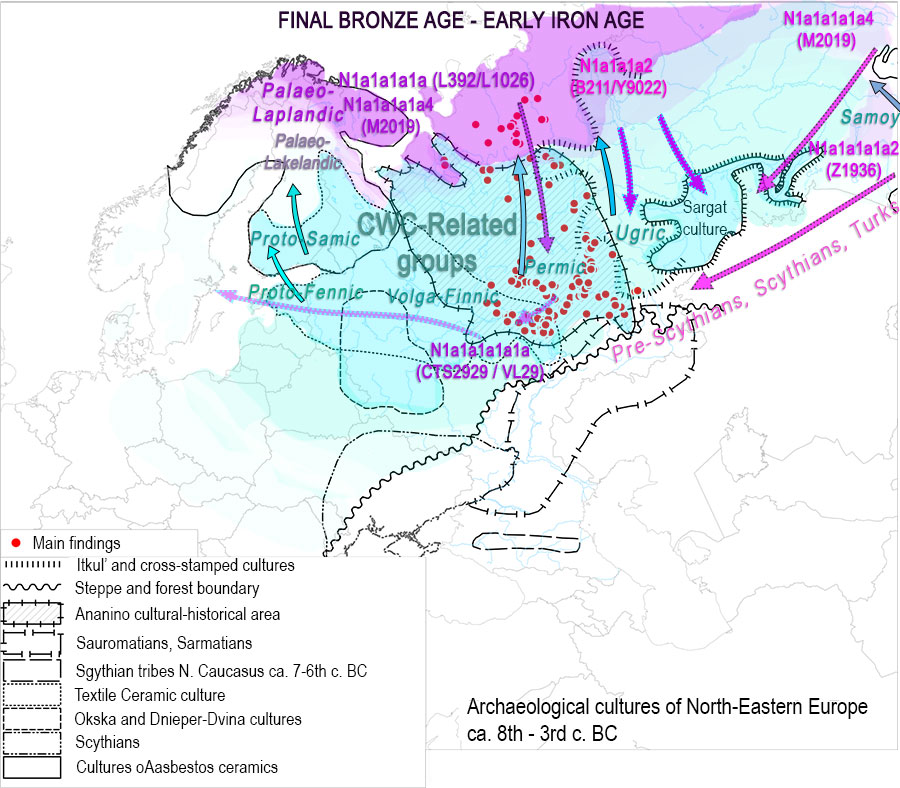
EDIT (2 APR): Interesting is the specific subclade to which ancient Mongolic-speaking Avars belong (information from Yfull) N1c-F4205 (TMRCA ca. 500 BC), subclade of N1c-Y6058 (formed ca. 2800 BC, TMRCA ca. 2800 BC). This branch also gives the “European” branch N1c-CTS10760 (formed ca. 2800 BC, TMRCA ca. 2100 BC), and is subclade of a branch of N1c-L392 (formed ca. 4400 BC, TMRCA ca. 2800 BC). A northern expansion of N1c-L392 is probably represented by its branch N1c-Z1936 (formed ca. 2800, TMRCA ca. 2100 BC), the most likely candidate to appear in the Kola Peninsula in the Bronze Age as the Palaeo-Laplandic population (see here). Read more about potential routes of expansion of haplogroup N.
On the other hand, R1a-Z280 lineages form a tight cluster connecting Permic with Ugric groups, with R1a-Z93 showing early isolation (probably) between Cis-Urals and Trans-Urals regions. While both Corded Ware lineages in Finno-Ugrians are most likely related to the Abashevo expansion through Seima-Turbino and the Andronovo-like Horizon (and potentially later Eurasian expansions), a plausible hypothesis would be that Finno-Ugrians are related to an expansion of R1a-Z283 haplogroups (we already knew about the Finno-Permic connection), while the ancient connection between Permians and Hungarians with R1a-Z93 would correspond to this haplogroup’s potentially tighter link with an early Samoyedic split.
I don’t think that an explosive expansion of eastern Corded Ware groups of R1a-Z645 lineages will show a clear-cut division of haplogroups among Eastern Uralic groups, though, and culturally I doubt we will have such a clear image, either (similar to how the explosive expansion of Bell Beakers cannot be easily divided by regional/language group into R1b-L151 subclades before the known bottlenecks). Relevant in this regard are the known Z93 samples from the Árpád dynasty.
Nevertheless, this data may represent a slightly more recent wave of R1a-Z280 lineages linked to the expansion of Ugric into the Trans-Uralian region, after their split from Finno-Permic, still in close contact with Indo-Iranians in Poltavka and Sintashta-Potapovka, evident from the early and late Indo-Iranian borrowings, during a common period when Samoyedic had already separated.
Such a “Z283 over Z93” layer in the Trans-Urals (and Cis-Urals?) forest-steppes would be similar to the apparent replacement of Z284 by Z282 in the Eastern Baltic during the Bronze Age (possibly with the second or Estonian Battle Axe wave or, much more likely during later population movements). Such an early R1a-Z93 split could potentially be supported also by the separation into bottlenecks under “Northern” (R1a-Z283) Finno-Ugric-speaking Abashevo-related groups and “Southern” (R1a-Z93) acculturated Indo-Iranian-speaking Abashevo migrants developing Sintashta-Potapovka admixing with Poltavka R1b-Z2103 herders.
Conclusion
Let’s review some of the most common myths about Hungarians (and Finno-Ugrians in general) repeated ad nauseam, side by side with my assertions:
❌ N (especially N1c-Tat) in ancient and modern samples represent the True Uralic™ N1c peoples including Magyar tribes? Nope.
✅ Ancient N (especially N1c-Tat) lineages among Uralic populations expanded relatively recently, and differently in different regions (including eastern steppe nomads and northern arctic populations) not associated with a particular language or language group? Yep (read the series on Corded Ware = Uralic expansion).
❌ Modern Hungarian R1a-Z280 lineages represent the majority of the native population, poor Slavic ‘peasants’ from the Carpathian Basin, forcibly acculturated by a minority of bad bad Hungarian hordes? Nope.
✅ Modern Hungarian R1a-Z280 subclades represent Ugric lineages in common with ancient R1a-Z645 Finno-Ugric populations from north-eastern Europe and the Trans-Urals? Yep (see Avars and Ugrians).
❌ Modern Hungarian R1a-Z93 lineages represent acculturated Iranian/Turkic peoples from the steppes? Not likely.
✅ Modern Hungarian R1a-Z93 lineages represent a remnant of the expansion of Corded Ware to the east, potentially more clearly associated with Samoyedic? Much more likely.
Sooo, the theory of a “diluted” Y-DNA in Modern Hungarians from originally fully N-dominated conquerors subjugating native R1a-Z280 Slavs from the Carpathian Basin is not backed up by genetic studies? The ethnic Iranian-Turkic R1a-Z93 federation in the steppes that ended up speaking Magyar is not real?? Who would’ve thunk.
Another true story whose rejection in genetics could not be predicted, like, not at all.
Totally unexpected, too, the drift of “R1a=IE” fans with the newest genetic findings towards a Molgen-like “Yamna/R1b = Vasconic-Caucasian”, “N1c = Uralic-Altaic”, and “R1a = the origin of the white world in Mother Russia”. So much for the supposed interest in “Steppe ancestry” and fancy statistics.
Related
- Mitogenomes from Avar nomadic elite show Inner Asian origin
- The complex origin of Samoyedic-speaking populations
- Corded Ware—Uralic (IV): Hg R1a and N in Finno-Ugric and Samoyedic expansions
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- The traditional multilingualism of Siberian populations
- Iron Age bottleneck of the Proto-Fennic population in Estonia
- Y-DNA haplogroups of Tuvinian tribes show little effect of the Mongol expansion
- Corded Ware—Uralic (I): Differences and similarities with Yamna
- Haplogroup R1a and CWC ancestry predominate in Fennic, Ugric, and Samoyedic groups
- On the origin and spread of haplogroup R1a-Z645 from eastern Europe
- Oldest N1c1a1a-L392 samples and Siberian ancestry in Bronze Age Fennoscandia