The Nightmare Week of “N1c=Uralic” proponents (see here) continues, now with preprint Y-chromosome haplogroups from Hun, Avar and conquering Hungarian period nomadic people of the Carpathian Basin, by Neparaczki et al. bioRxiv (2019).
Abstract:
Hun, Avar and conquering Hungarian nomadic groups arrived into the Carpathian Basin from the Eurasian Steppes and significantly influenced its political and ethnical landscape. In order to shed light on the genetic affinity of above groups we have determined Y chromosomal haplogroups and autosomal loci, from 49 individuals, supposed to represent military leaders. Haplogroups from the Hun-age are consistent with Xiongnu ancestry of European Huns. Most of the Avar-age individuals carry east Eurasian Y haplogroups typical for modern north-eastern Siberian and Buryat populations and their autosomal loci indicate mostly unmixed Asian characteristics. In contrast the conquering Hungarians seem to be a recently assembled population incorporating pure European, Asian and admixed components. Their heterogeneous paternal and maternal lineages indicate similar phylogeographic origin of males and females, derived from Central-Inner Asian and European Pontic Steppe sources. Composition of conquering Hungarian paternal lineages is very similar to that of Baskhirs, supporting historical sources that report identity of the two groups.
Interesting excerpts (emphasis mine):
All N-Hg-s identified in the Avars and Conquerors belonged to N1a1a-M178. We have tested 7 subclades of M178; N1a1a2-B187, N1a1a1a2-B211, N1a1a1a1a3-B197, N1a1a1a1a4-M2118, N1a1a1a1a1a-VL29, N1a1a1a1a2-Z1936 and the N1a1a1a1a2a1c1-L1034 subbranch of Z1936. The European subclades VL29 and Z1936 could be excluded in most cases, while the rest of the subclades are prevalent in Siberia 23 from where this Hg dispersed in a counter-clockwise migratory route to Europe (…). All the 5 other Avar samples belonged to N1a1a1a1a3-B197, which is most prevalent in Chukchi, Buryats, Eskimos, Koryaks and appears among Tuvans and Mongols with lower frequency.
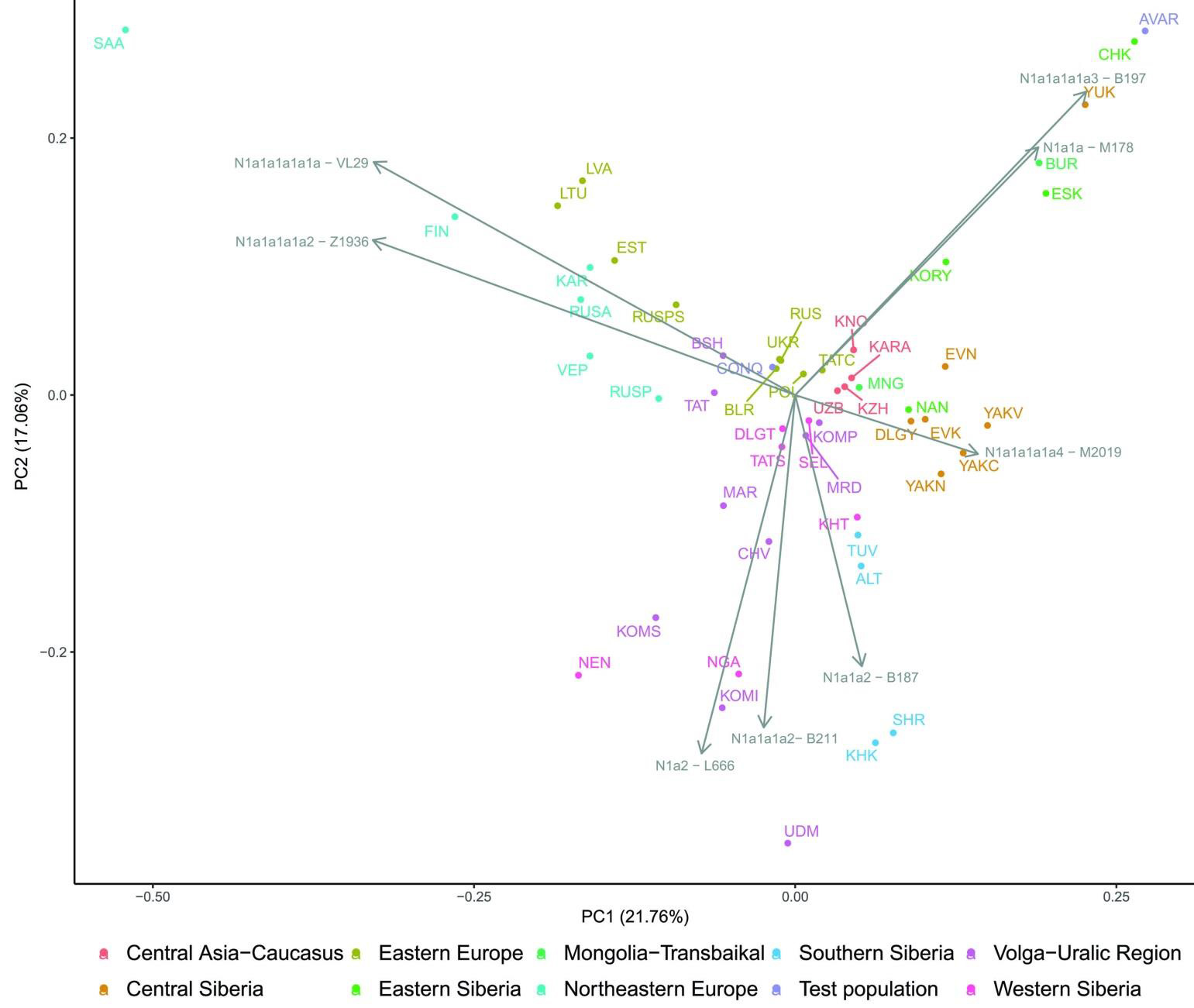
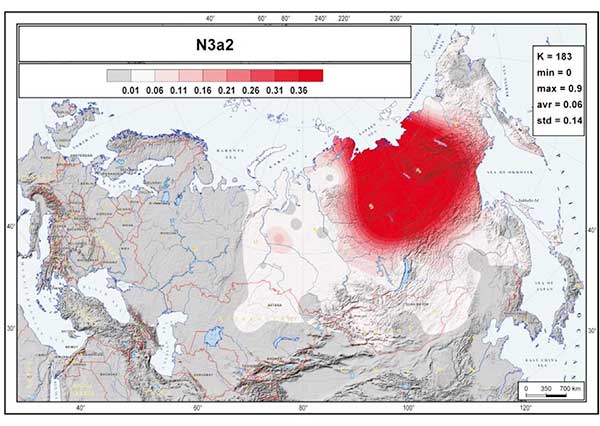
By contrast two Conquerors belonged to N1a1a1a1a4-M2118, the Y lineage of nearly all Yakut males, being also frequent in Evenks, Evens and occurring with lower frequency among Khantys, Mansis and Kazakhs.
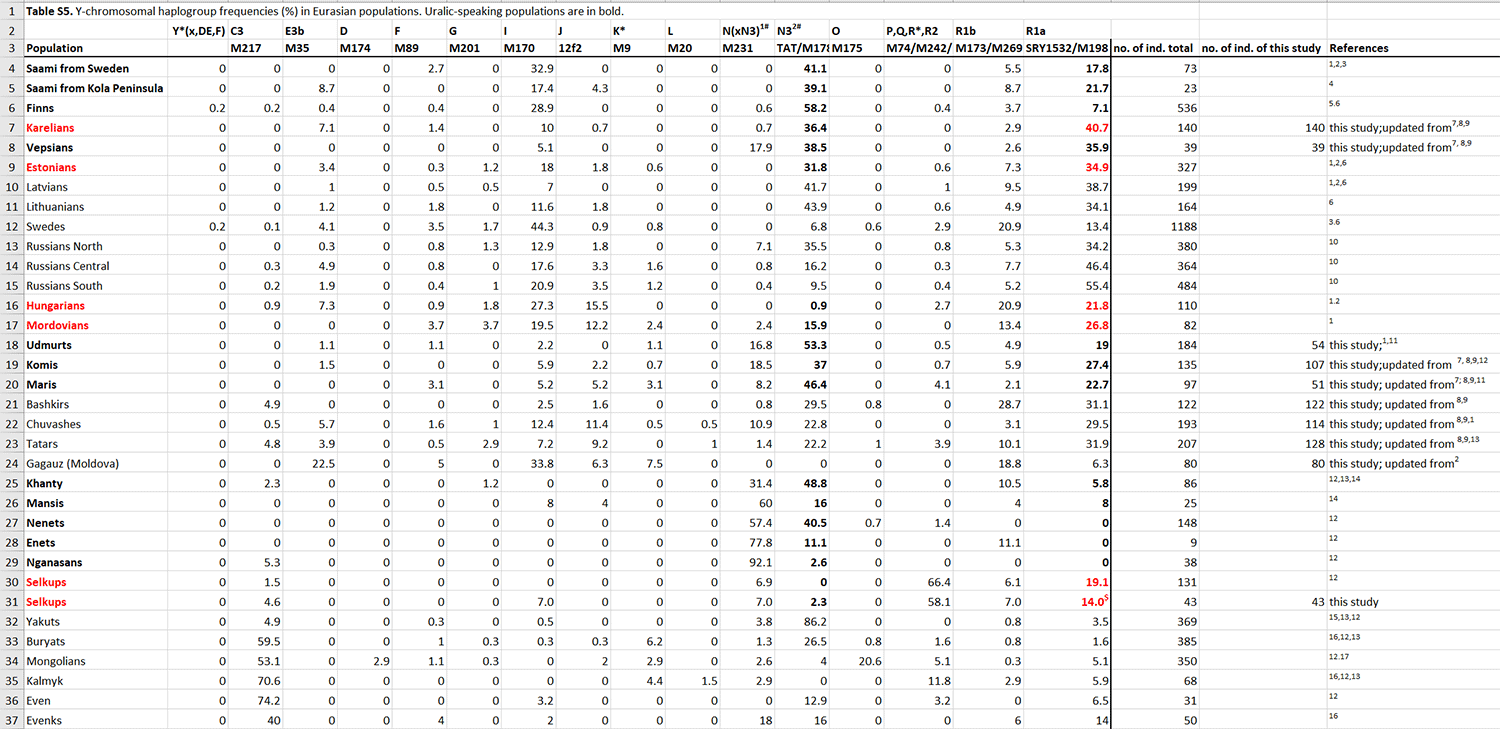
Three Conqueror samples belonged to Hg N1a1a1a1a2-Z1936 , the Finno-Permic N1a branch, being most frequent among northeastern European Saami, Finns, Karelians, as well as Komis, Volga Tatars and Bashkirs of the Volga-Ural region.Nevertheless this Hg is also present with lower frequency among Karanogays, Siberian Nenets, Khantys, Mansis, Dolgans, Nganasans, and Siberian Tatars.
The west Eurasian R1a1a1b1a2b-CTS1211 subclade of R1a is most frequent in Eastern Europe especially among Slavic people. This Hg was detected just in the Conqueror group (K2/18, K2/41 and K1/10). Though CTS1211 was not covered in K2/36 but it may also belong to this sub-branch of Z283.
Hg I2a1a2b-L621 was present in 5 Conqueror samples, and a 6th sample form Magyarhomorog (MH/9) most likely also belongs here, as MH/9 is a likely kin of MH/16 (see below). This Hg of European origin is most prominent in the Balkans and Eastern Europe, especially among Slavic speaking groups. It might have been a major lineage of the Cucuteni-Trypillian culture and it was present in the Baden culture of the Chalcolithic Carpathian Basin.
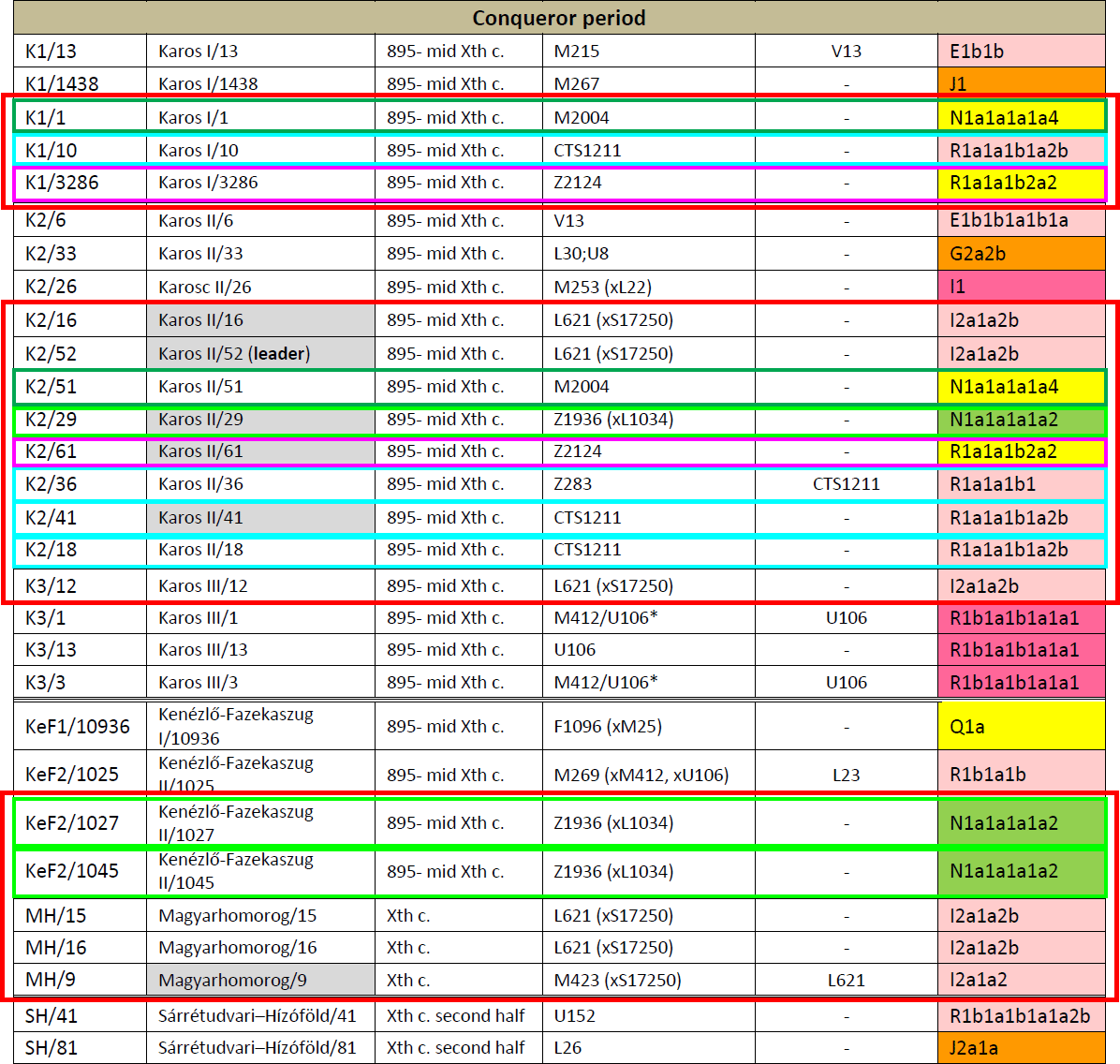
We identified potential relatives within Conqueror cemeteries but not between them. The uniform paternal lineages of the small Karos3 (19 graves) and Magyarhomorog (17 graves) cemeteries approve patrilinear organization of these communities. The identical I2a1a2b Hg-s of Magyarhomorog individuals appears to be frequent among high-ranking Conquerors, as the most distinguished graves in the Karos2 and 3 cemeteries also belong to this lineage. The Karos2 and Karos3 leaders were brothers with identical mitogenomes 11 and Y-chromosomal STR profiles (Fóthi unpublished). The Sárrétudvari commoner cemetery seems distinct from the others, containing other sorts of European Hg-s. Available Y-chromosomal and mtDNA data from this cemetery suggest that common people of the 10th century rather represented resident population than newcomers. The great diversity of Y Hg-s, mtDNA Hg-s, phenotypes and predicted biogeographic classifications of the Conquerors indicate that they were relatively recently associated from very diverse populations.
Surprising about the Hungarian conquerors – although in line with the historical accounts – is the varied patrilineal origin of clans, including Q1a, G2a2b, I1, E1b1b, R1b, J1, or J2 – some of which (depending on specific lineages) may have appeared earlier in the Carpathian Basin or south-eastern Europe.
However, out of the 27 conqueror elite samples, 17 are of haplogroups most likely related to Ugric populations beyond the Urals: R1a-Z645, I2-L621, and two specific N1a-L392 lineages (see below). In fact, there are three high-ranking conqueror elites of hg. I2-L621 (one of them termed a “leader”, brother to an unpublished leader of Karos3, and all of them possibly family), one of hg. R1a-Z280, one of hg. R1a-Z93 (which should be added to the Árpáds), and one of hg. N1a-Z1936, which gives a good idea of the ruling class among the elite Ugric settlers.
NOTE. The Q1a sample is also likely to be found in the mixed population of the West Siberian forest-steppes, since it was found in Mesolithic-Neolithic samples from eastern Europe to Lake Baikal, and in Bronze Age Siberian groups, although admittedly it may have formed part of an Avar Transtisza group, or even earlier Hunnic or Scythian groups along the steppes. Without precise subclades it’s impossible to know.

I2a-L621
I2a-L621 (xS17250) or I2a1b2 in the old nomenclature, is found in 6 early conquerors (including one leader), on a par with R1a and N samples. This haplogroup is found widely distributed in ancient samples, due to its early split (formed ca. 9200 BC, TMRCA ca. 4500 BC) and expansion, probably with Neolithic populations. I can’t seem to find samples of this early haplogroup from the Carpathian Basin, as mentioned in the text, although it wouldn’t be strange, because it appears also in Neolithic Iberia, and in modern populations from western Europe.
Nevertheless, I2a-L621 samples seem to be concentrated mainly in Mesolithic-Neolithic cultures of Fennoscandia, and appeared also in Sikora et al. (2017) in a sample of the High Middle Ages from Sunghir (ca. AD 1100-1200), probably from the Vladimir-Suzdalian Rus’, in a region where clearly tribes of Volga Finns were being assimilated at the time. The reported SNP call by Genetiker is A16681 (see Yfull), deep within I2a-CTS10228. It is possibly also behind a modern Saami from Chalmny Varre (ca. AD 1800) of hg. I2a in Lamnidis et al. (2018).
Lacking precise subclades from Hungarian conquerors this is pure speculation, but modern samples may also point to I2a-CTS10228 (formed ca. 3100 BC, TMRCA ca. 1800 BC) as a Finno-Ugric lineage in common with R1a, which must have expanded to the Urals and beyond with eastern Corded Ware groups or (more likely) succeeding cultures. This is in line with the association of certain I2a lineages with modern Uralic peoples or populations from their historical regions in eastern Europe, and linked thus to the most likely homeland of Uralians in the eastern European forests:
R1a-Z645
Regarding the important question of the ethnic makeup of Ugric populations stemming from the Urals, the most interesting (and expected) data is the presence of R1a-Z645 lineages among high-ranking conquerors, in particular four R1a-Z280 subclades proper of Finno-Ugrians.
This proves that, in line with the old split and expansion of R1a-CTS1211 (formed ca. 2600 BC, TMRCA ca. 2400 BC), and its finding in Bronze Age Fennoscandian samples, only some late R1a-Z280 (xZ92) lineages (see Z280 on YFull) may show a clear identification with early acculturated Uralic speakers, with the main early acculturated Balto-Slavic R1a haplogroup remaining R1a-M458.
I recently hypothesized this late connection of Slavs with very specific R1a-Z280 (xZ92) lineages based on analyses of modern populations (like Slovenians), because the connection of ancient Finno-Ugrians with modern Z92 samples was already evident:
(…) subclades of hg. R1a1a1b1a2-Z280 (xR1a1a1b1a2a-Z92) seem to have also been involved in early Slavic expansions, like R1a1a1b1a2b3a-CTS3402 (formed ca. 2200 BC, TMRCA ca. 2200 BC), found among modern West, South, and East Slavic populations and in Fennoscandia, prevalent e.g. among modern Slovenians which points to a northern origin of its expansion (Maisano Delser et al. 2018).
This finding also supports the expected shared R1a-Z280 lineages among ancient Finno-Ugric populations, as predicted from the study of modern Permic and Ugric peoples in Dudás et al. (2019).
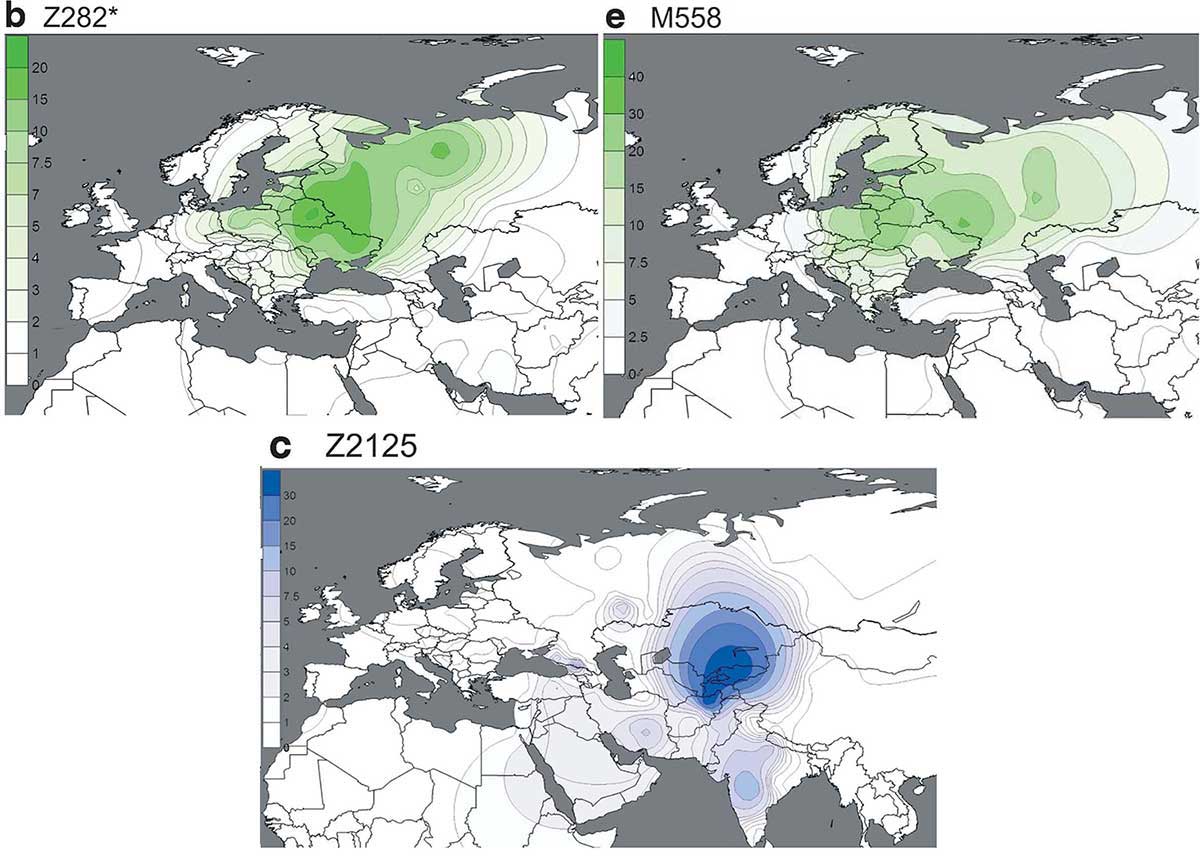
Furthermore, while we don’t have precise R1a-Z93 lineages to compare with the new Hunnic sample reported, we already know that some archaic R1a-Z2124 subclades stem from the forest-steppe areas of the Cis- and Trans-Urals, and the two newly reported R1a-Z93 Hungarian conqueror elites, like those of the Árpád dynasty, probably belong to them.
There is an obvious lack of continuity in specific paternal lineages among the Hunnic, the Avar, and the Conqueror periods, which makes any simplistic identification of all R1a-Z93 lineages as stemming from Avars, Huns, or the Iron Age Pontic-Caspian steppes clearly flawed. Comparing R1a-Z93 in Hungarian Conquerors with Huns is like comparing them with samples of the Srubna or earlier periods… Similarly, comparing the Hunnic R1b-U106 or the early Avar I1 to later Hungarian samples is not warranted without precise subclades, because they most likely correspond to different Germanic populations: Goths among Huns, then Longobards, then likely peoples descended from Franks and Irish Monks (the latter with R1b-P312).
N1a-L392
Second behind R1a subclades are, as expected, N1a-L392 (N1c in the old nomenclature).
Avars are dominated by a specific N1a-L392 subclade, N1a-B197, as we recently discovered in Csáky et al. (2019).
Hungarian conquerors show three N1a-Z1936 subclades, which is known to stem from the northern Ural region, including the Arctic (likely Palaeo-Laplandic peoples) and cross-stamped cultures of the northern Eurasian forests.
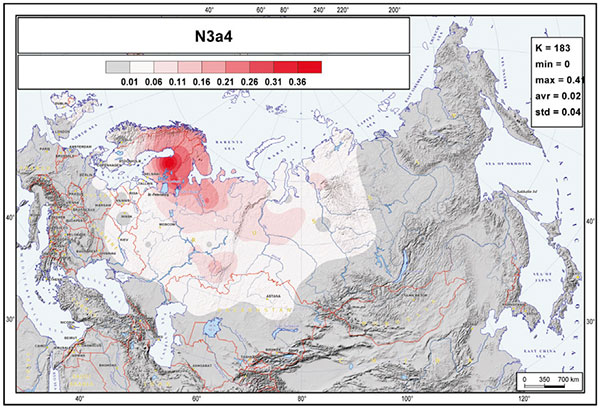
On the other hand, the two N1a-M2118 lineages are more clearly associated with Palaeo-Siberian populations east of the Urals, but became incorporated into the Ugric stock in the Trans-Urals region probably in the same way as N1a-Z1936, by infiltration from (and acculturation of) hunter-gatherers of forest and taiga cultures.
NOTE. You can read more about the infiltration of N1a lineages in the recent post Corded Ware—Uralic (IV): Hg R1a and N in Finno-Ugric and Samoyedic expansions, and in the specific sections for each Uralic group in A Clash of Chiefs.
Conclusion
The picture offered by the paper on Hungarian Conquerors, while in line with historical accounts of multi-ethnic tribes incorporating regional lineages, shows nevertheless patrilineal clans clearly associated with Uralic peoples, in a distribution which could have been easily inferred from ancient Trans-Uralian forest-steppe cultures and modern samples (even regarding I2a-L621).
In spite of this, there is a great deal of discussion in the paper about specific N1a subclades in Hungarian conquerors, while the presence of R1a-Z280 (among early Magyar elites!) is interpreted, as always, as recently acculturated Slavs. This is sadly coupled with the simplistic identification of I2a-L621 as of local origin around the Carpathians.
The introduction of the paper to the history of Hungarians is also weird, for example giving credibility to the mythic accounts of the Árpád dynasty’s origin in Attila, which is in line, I guess, with what the authors intended to support all along, i.e. the association of Magyars with Turks from the Eurasian steppes, which they are apparently willing to achieve by relating them to haplogroup R1a-Z93…
The conclusion is thus written to appease modern nation-building myths more than anything else, like many other papers before it:
It is generally accepted that the Hungarian language was brought to the Carpathian Basin by the Conquerors. Uralic speaking populations are characterized by a high frequency of Y-Hg N, which have often been interpreted as a genetic signal of shared ancestry. Indeed, recently a distinct shared ancestry component of likely Siberian origin was identified at the genomic level in these populations, modern Hungarians being a puzzling exception36. The Conqueror elite had a significant proportion of N Hgs, 7% of them carrying N1a1a1a1a4-M2118 and 10% N1a1a1a1a2-Z1936, both of which are present in Ugric speaking Khantys and Mansis. At the same time none of the examined Conquerors belonged to the L1034 subclade of Z1936, while all of the Khanty Z1936 lineages reported in 37 proved to be L1034 which has not been tested in the 23 study. Population genetic data rather position the Conqueror elite among Turkic groups, Bashkirs and Volga Tatars, in agreement with contemporary historical accounts which denominated the Conquerors as “Turks”. This does not exclude the possibility that the Hungarian language could also have been present in the obviously very heterogeneous, probably multiethnic Conqueror tribal alliance.
So, back to square one, and new circular reasoning: If ancient populations from north-eastern Europe believed to represent ancient Finno-Ugrians are of R1a-Z645 lineages, it’s because they were not Finno-Ugric speakers. If ancient and modern populations known to be of Finno-Ugric language show clear connections with R1a-Z645, it’s because they are “multi-ethnic”.
The only stable basis for discussion in genetic papers, apparently, is the own making of geneticists, with their traditional 2000s “R1a=Indo-European” and “N1c=Uralic”, coupled with national beliefs. It does not matter how many predictions based on that have been proven wrong, or how many predictions based on the Corded Ware = Uralic expansion have been proven right.
Related
- R1a-Z280 and R1a-Z93 shared by ancient Finno-Ugric populations; N1c-Tat expanded with Micro-Altaic
- Scytho-Siberians of Aldy-Bel and Sagly, of haplogroup R1a-Z93, Q1b-L54, and N
- Mitogenomes from Avar nomadic elite show Inner Asian origin
- The complex origin of Samoyedic-speaking populations
- Corded Ware—Uralic (IV): Hg R1a and N in Finno-Ugric and Samoyedic expansions
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- The traditional multilingualism of Siberian populations
- Iron Age bottleneck of the Proto-Fennic population in Estonia
- Y-DNA haplogroups of Tuvinian tribes show little effect of the Mongol expansion
- Corded Ware—Uralic (I): Differences and similarities with Yamna
- Haplogroup R1a and CWC ancestry predominate in Fennic, Ugric, and Samoyedic groups
- On the origin and spread of haplogroup R1a-Z645 from eastern Europe
- Oldest N1c1a1a-L392 samples and Siberian ancestry in Bronze Age Fennoscandia