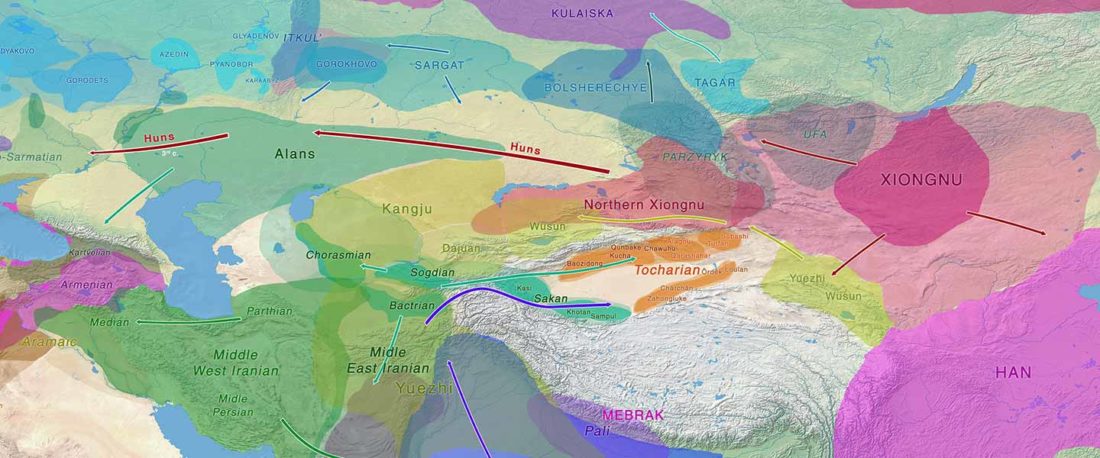
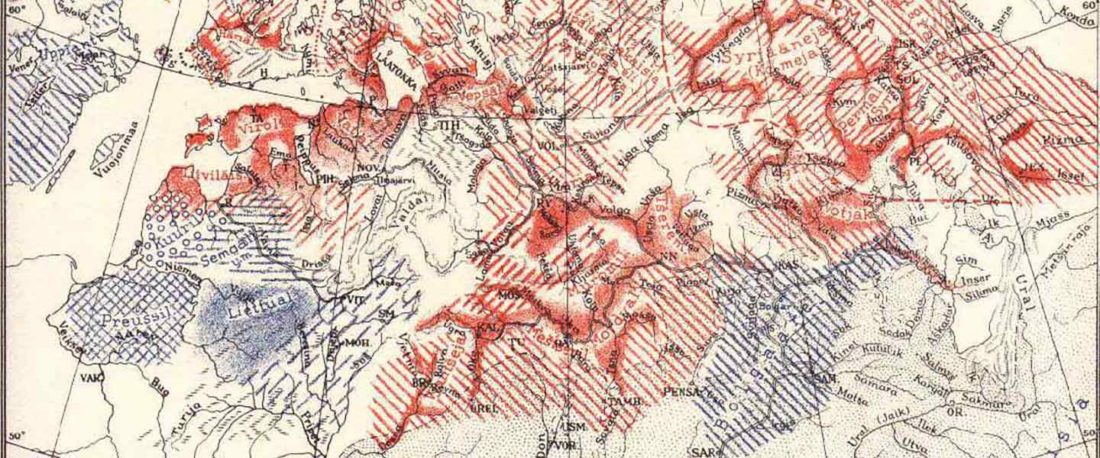
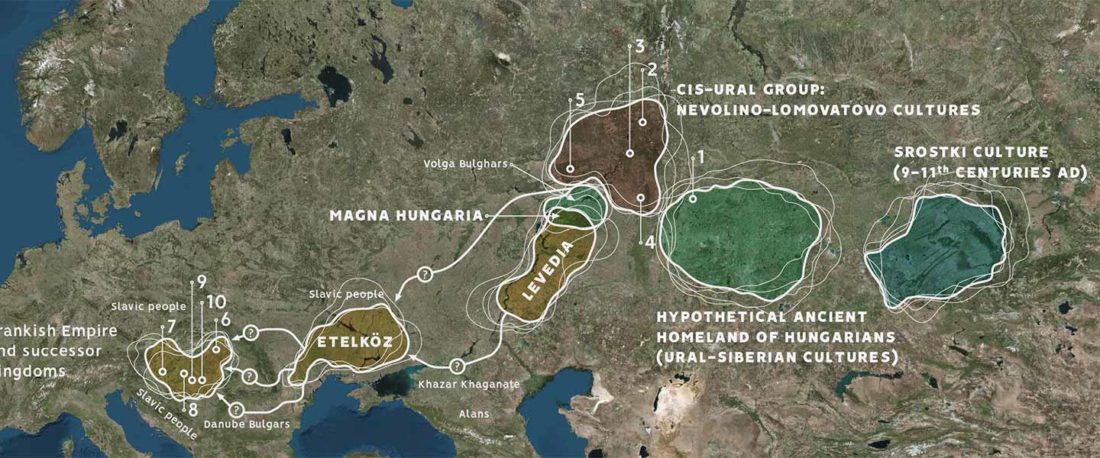
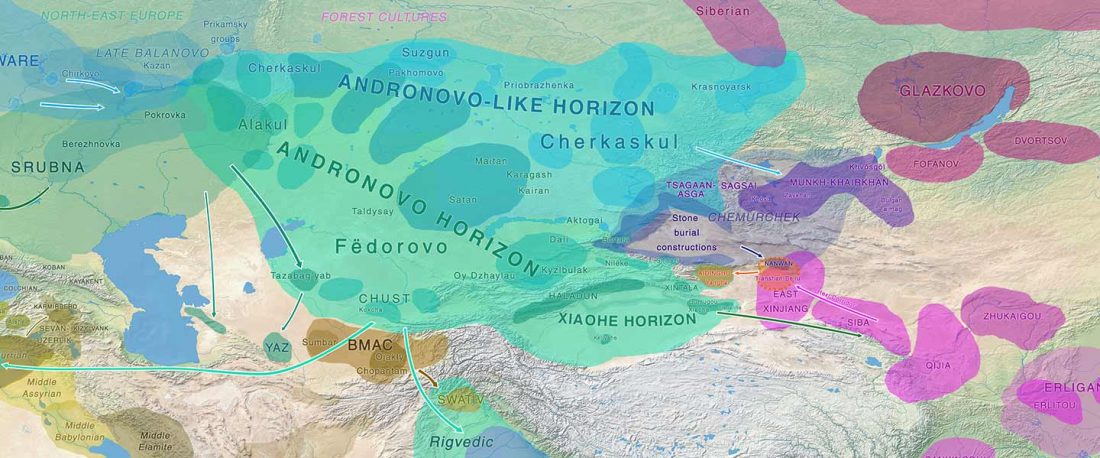
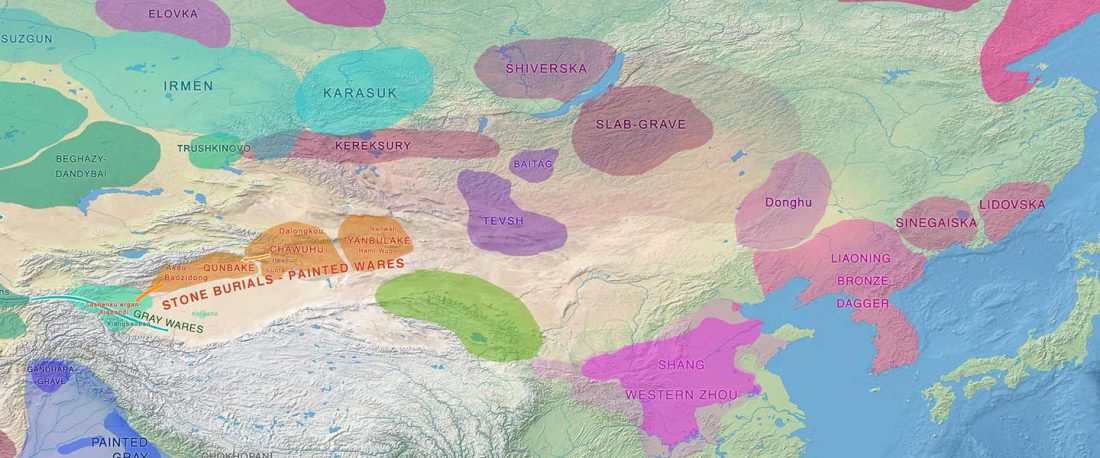
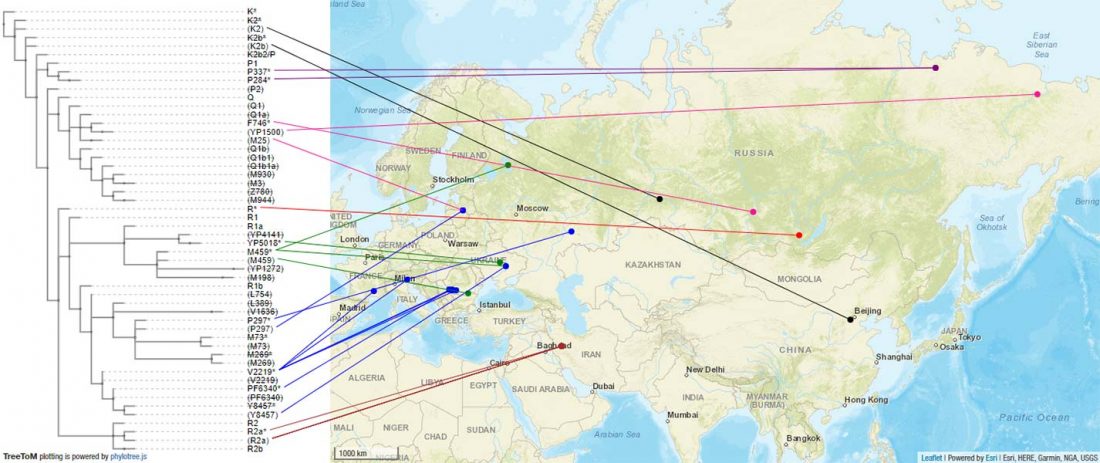
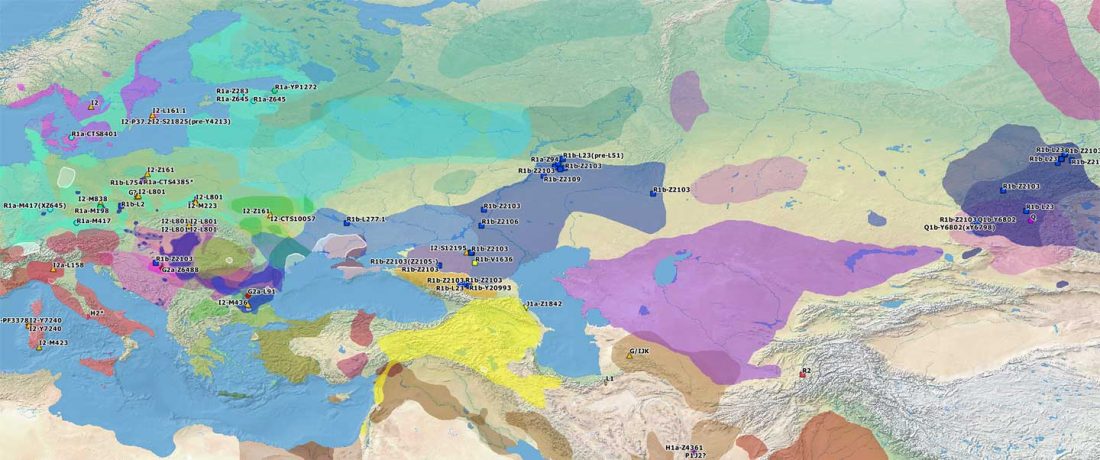
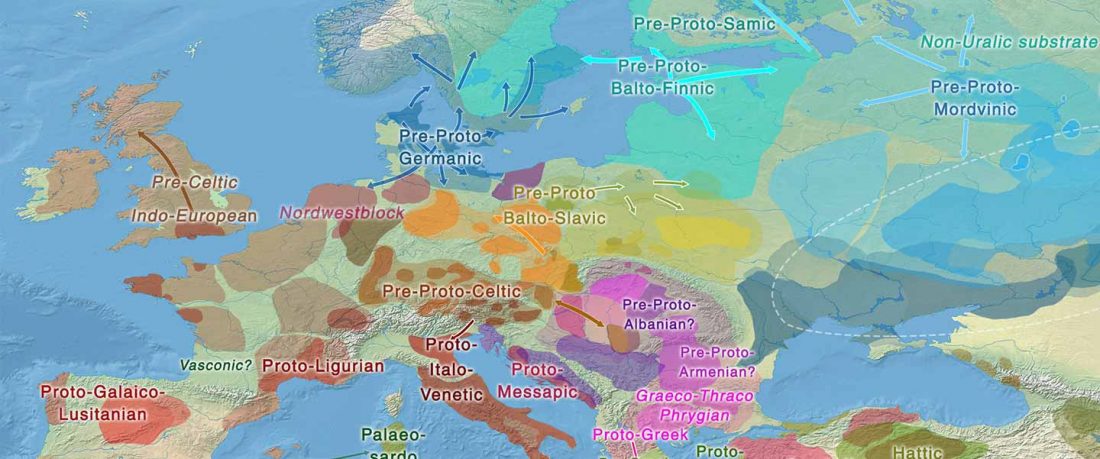
Recent paper (behind paywall) Genetic evidence suggests a sense of family, parity and conquest in the Xiongnu Iron Age nomads of Mongolia, by Keyser, Zvénigorosky, Gonzalez, et al. Human Genetics (2020).
Interesting excerpts (emphasis mine):
Site and bodies
… Read the rest “Xiongnu Y-DNA connects Huns & Avars to Scytho-Siberians”The Tamir Ulaan Khoshuu (TUK) cemetery is located near the confluence of the Tamir River and the Orkhon River in the Arkhangai Aimag (Central Mongolia), about four hundred kilometers west of the capital of Mongolia, Ulaanbaatar. It encompasses an area of 22 hectares located on a prominent granitic outcrop and comprises a total of 397 graves, delimited by stone circles. (…)