Open access paper New genetic evidence of affinities and discontinuities between bronze age Siberian populations, by Hollard et al., Am J Phys Anthropol. (2018) 00:1–11.
NOTE. This seems to be a peer-reviewed paper based on a more precise re-examination of the samples from Hollard’s PhD thesis, Peuplement du sud de la Sibérie et de l’Altaï à l’âge du Bronze : apport de la paléogénétique (2014).
Interesting excerpts:
Afanasevo and Yamna
The Afanasievo culture is the earliest known archaeological culture of southern Siberia, occupying the Minusinsk-Altai region during the Eneolithic era 3600/3300 BC to 2500 BC (Svyatko et al., 2009; Vadetskaya et al., 2014). Archeological data showed that the Afanasievo culture had strong affinities with the Yamnaya and pre-Yamnaya Eneolithic cultures in the West (Grushin et al., 2009). This suggests a Yamnaya migration into western Altai and into Afanasievo. Note that, in most current publications, “the Yamnaya culture” combines the so-called “classical Yamnaya culture” of the Early Bronze Age and archeological sites of the preceding Repin culture in the middle reaches of the Don and Volga rivers. In the present article we conventionally use the term Yamnaya in the same sense, in which case the beginning of the “Yamnaya culture” can be dated after the middle of the 4th millennium BC, when the Afanasievo culture appeared in the Altai.
Because of numerous traits attributed to early Indo-Europeans and cultural relations with Kurgan steppe cultures, members of the Afanasievo culture are believed to have been Indo-European speakers (Mallory and Mair, 2000). In a recent whole-genome sequencing study, Allentoft et al. (2015) concluded that Eastern Yamnaya individuals and Afanasievo individuals were genetically indistinguishable. Moreover, this study and one published concurrently by Haak et al. (2015) analyzed 11 Eastern Yamnaya males and showed that all of them belonged to the R1b1a1a (formerly R1b1a) (…)
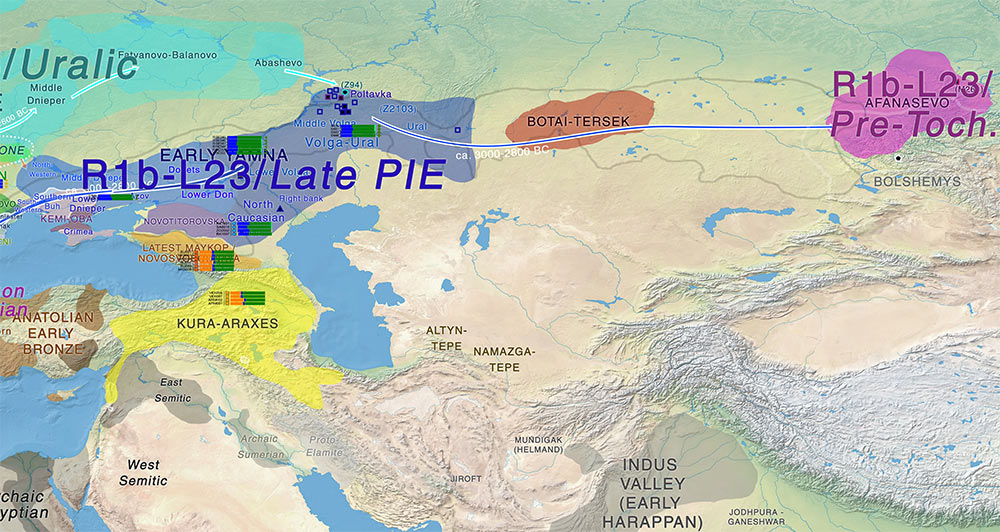
Published works indicate that R1b was a predominant haplogroup from the late Neolithic to the early Bronze Age, notably in the Bell Beaker and Yamnaya cultures (Allentoft et al., 2015; Haak et al., 2015; Lee et al., 2012; Mathieson et al., 2015). Nearly 100% of the Afanasievo men we typed belonged to the R1b1a1a subhaplogroup and, for at least three of them, more precisely to the L23 (xM412) subclade. (…)
(…) our results therefore support the hypothesis of a genetic link between Afanasievo and Yamnaya. This also suggests that R1b was indeed dominant in the early Bronze Age Siberian steppe, at least in individuals that were buried in kurgans (possibly an elite part of the population). The geographical and temporal distribution of subhaplogroup R1b1a1a supports the hypothesis of population expansion from West to East in the Eurasian steppe during this period. It should however be noted that the Yamnaya burials from which the samples for DNA analysis were obtained (Allentoft et al., 2015; Haak et al., 2015; Mathieson et al., 2015) were dated within the limits of the Afanasievo period. Ancestors of both East Yamnaya and Afanasievo populations must therefore be sought in the context of earlier Eneolithic cultures in Eastern Europe. Sufficient Y-chromosomal data from such Eneolithic populations is, unfortunately, not yet available.
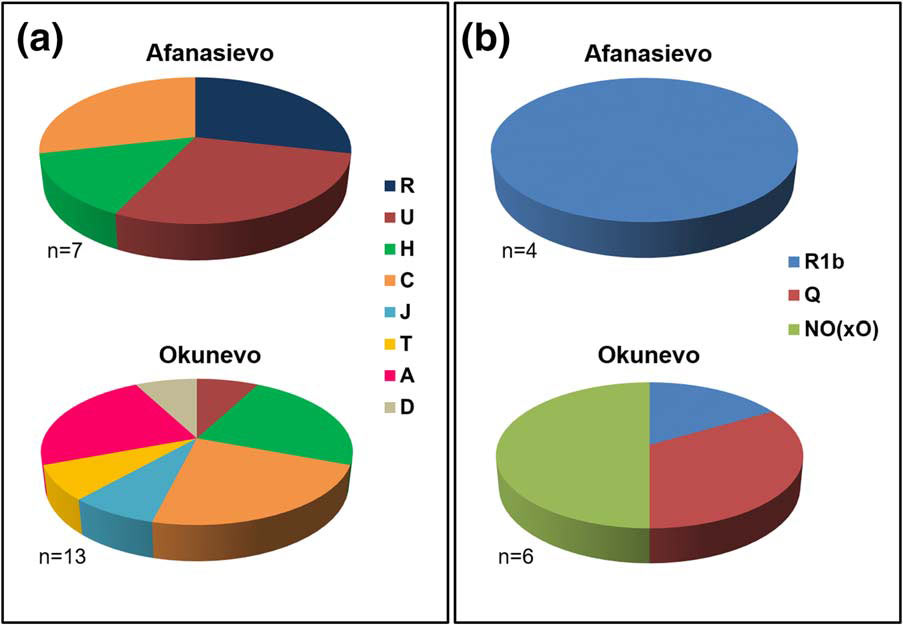
Okunevo and paternal lineage shift in South Siberia
Results obtained in the current study, from more than a dozen Okunevo individuals belonging to the earliest stage of Okunevo culture, that is the Uibat period (2500–2200 BC) (Lazaretov, 1997), suggest a discontinuity in the genetic pool between Afanasievo and Okunevo cultures. Although Y-chromosomal data obtained for bearers of the Okunevo culture showed that one individual carried haplogroup R1b, most Okunevo Y-haplogroups are representative of an Asian component represented by paternal lineages Q and NO1.
Okunevo carrier of Y-haplogroup Q1b1a-L54, which also supports this hypothesis (L54 being a marker of the lineage from which M3, the main Ameridian lineage, arose). Okunevo people could therefore be a remnant paleo-Siberian population with possible Afanasievo input, as suggested by the presence of the R1b1a1a2a subhaplogroup in one individual.
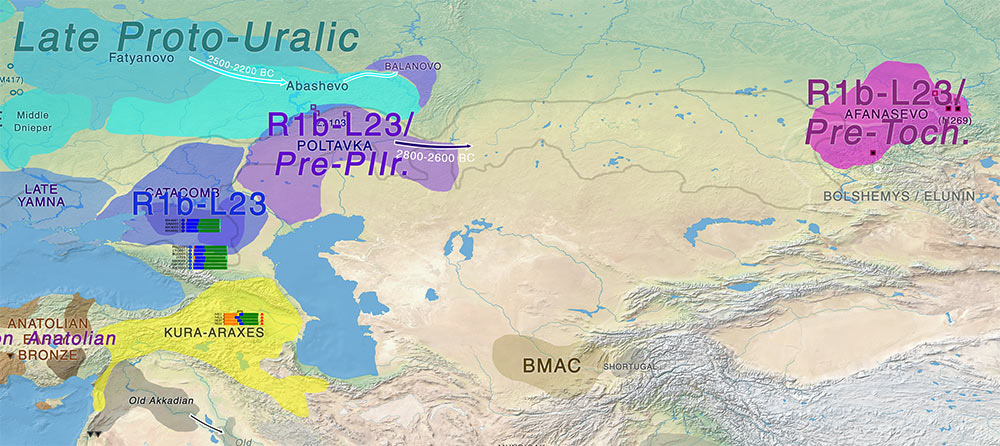
Replacement of Asian Indo-European elite lineages by R1a
Published genetic data from the late Bronze Age Andronovo culture from the Minusinsk Basin (Keyser et al., 2009), the Sintashta culture from Russia (Allentoft et al., 2015) and the Srubnaya culture from the region of Samara (Mathieson et al., 2015), show that males did not belong to Y-haplogroup R1b but mostly to R1a clades: there appears to have been a change in the dominant Y-chromosomal haplogroup between the early and the late Bronze Age in these regions. Moreover, as described in Allentoft et al. (2015), the Andronovo and Sintashta peoples were closely related to each other but clearly distinct from both Yamnaya and Afanasievo. Although these results do not imply that Y-haplogroup R1b was entirely absent in these later populations, they could correspond to a replacement of the elite between these two main periods and therefore a difference in the haplogroups of the men that were preferentially buried.
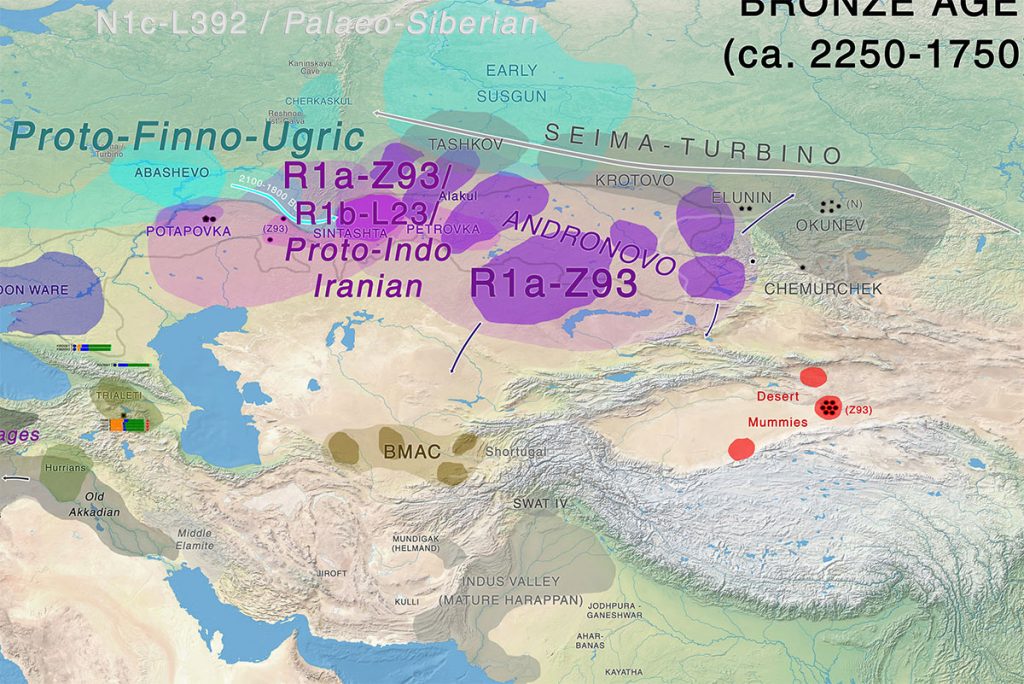
Afanasevo and the Tarim Basin
The discovery, in the Tarim Basin, of well-preserved mummies from the Bronze Age allows for the construction of two hypotheses regarding the peopling of the Xinjiang province at this period. The “steppe hypothesis,” argues for a link with nomadic steppe herders (Hemphill and Mallory, 2004), possibly represented in this case by Afanasievo populations and their descendants (Mallory and Mair, 2000). However, newly published cultural data from the burial grounds of Gumugou (Wang, 2014) and Xiaohe (Xinjiang, 2003, 2007) shows material culture and burial rites incompatible with the Afanasievo culture. The earliest 14C date for Tarim Basin burials would place them at the turn of the 2nd millenium BC (Wang, 2013), 500 years after the Afanasievo period.
Instead, early Gumugou and Xiaohe burial grounds were contemporary with the start of the Andronovo period. Likewise, the Bronze Age population of the Xinjiang at Gumugou/Qäwrighul is not phenotypically closest to Afanasievo but to the Andronovo (Fedorovo) group of northeastern Kazakhstan and western Altai (Kozintsev, 2009). Our investigations demonstrate that Y-chromosomal lineage composition is also compatible with the notion that the ancient Tarim population was genetically distinct from the Afanasievo population. The only Y-haplogroup found by Li et al. (2010) in the Bronze Age Tarim Basin population was Y-haplogroup R1a, which suggests a proximity of this population with Andronovo groups rather than Afanasievo groups.
I don’t think these finds are much of a surprise based on what we already know, or need much explanation…
I would add that, once again, we have more proof that the movement of Okunevo and related ancient Siberian migrants from Central or North Asia will not be able to explain the presence of Uralic languages spread over North-East Europe and Scandinavia already during the Bronze Age.
Also interesting is to read in more peer-reviewed papers the idea of Late Indo-European speakers clearly linked to the expansion of patrilineally-related elite males marked by haplogroup R1b-L23, most likely since Eneolithic Khvalynsk/Repin cultures.
Related:
- North Asian mitogenomes hint at the arrival of pastoralists from West to East ca. 2800-1000 BC
- Hungarian mitogenomes similar to East and West Slavs, but genetic substratum predates their historic contacts
- Pre-Germanic born out of a Proto-Finnic substrate in Scandinavia
- Consequences of Damgaard et al. 2018 (III): Proto-Finno-Ugric & Proto-Indo-Iranian in the North Caspian region
- Consequences of Damgaard et al. 2018 (II): The late Khvalynsk migration waves with R1b-L23 lineages
- On Proto-Finnic language guesstimates, and its western homeland
- Early Indo-Iranian formed mainly by R1b-Z2103 and R1a-Z93, Corded Ware out of Late PIE-speaking migrations
- Oldest N1c1a1a-L392 samples and Siberian ancestry in Bronze Age Fennoscandia
- The concept of “Outlier” in Human Ancestry (III): Late Neolithic samples from the Baltic region and origins of the Corded Ware culture
- Genetic prehistory of the Baltic Sea region and Y-DNA: Corded Ware and R1a-Z645, Bronze Age and N1c
- Uralic as a Corded Ware substrate of Indo-Iranian, and loanwords in Finno-Ugric
- More evidence on the recent arrival of haplogroup N and gradual replacement of R1a lineages in North-Eastern Europe