Over the past week or so, since the publication of new Corded Ware samples in Narasimhan, Patterson et al. (2019) and after finding out that the R1a-M417 star-like phylogeny may have started ca. 3000 BC, I have been ruminating the relevance of contradictory data about the Ukraine_Eneolithic_o sample from Alexandria, its potential wrong radiocarbon date, and its implications for the Indo-European question.
How many other similar ‘controversial’ samples are there which we haven’t even considered? And what mechanisms are in place to control that the case of Hajji_Firuz_CA I2327 is not repeated?
Ukraine Eneolithic outlier I6561
It was not the first time that I (or many others) have alternatively questioned its subclade or its date, but the contradictory data seem to keep piling up. We can still explain all these discrepancies by assuming that the radiocarbon date is correct – seeing how it is a direct and newly reported lab analysis – because it is an isolated individual from a poorly sampled region, so he may actually be the first one to show features proper of later Corded Ware-related samples.
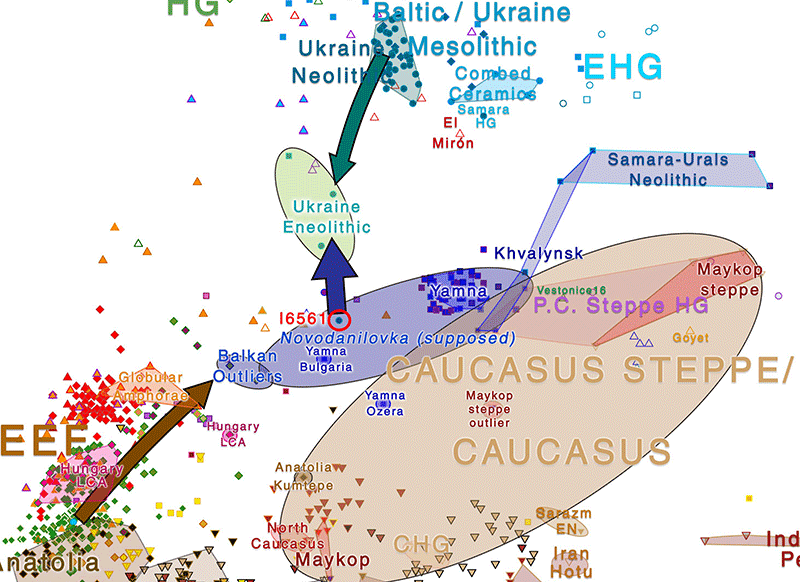
The individual seems to be especially relevant for the Indo-European and Uralic homeland question. The last one to mention this sample in a publication was Anthony (2019), who considered it in common with two other Eneolithic samples from Dereivka to show how Anatolian farmer-related ancestry first appeared in the recently opened CHG mating network of the Pontic-Caspian steppes and forest-steppes during the Middle Eneolithic, after the expansion of Khvalynsk:
The currently oldest sample with Anatolian Farmer ancestry in the steppes in an individual at Aleksandriya, a Sredni Stog cemetery on the Donets in eastern Ukraine. Sredni Stog has often been discussed as a possible Yamnaya ancestor in Ukraine (Anthony 2007: 239- 254). The single published grave is dated about 4000 BC (4045–3974 calBC/ 5215±20 BP/ PSUAMS-2832) and shows 20% Anatolian Farmer ancestry and 80% Khvalynsk-type steppe ancestry (CHG&EHG). His Y-chromosome haplogroup was R1a-Z93, similar to the later Sintashta culture and to South Asian Indo-Aryans, and he is the earliest known sample to show the genetic adaptation to lactase persistence (I3910-T). Another pre-Yamnaya grave with Anatolian Farmer ancestry was analyzed from the Dnieper valley at Dereivka, dated 3600-3400 BC (grave 73, 3634–3377 calBC/ 4725±25 BP/ UCIAMS-186349). She also had 20% Anatolian Farmer ancestry, but she showed less CHG than Aleksandriya and more Dereivka-1 ancestry, not surprising for a Dnieper valley sample, but also showing that the old fifth-millennium-type EHG/WHG Dnieper ancestry survived into the fourth millennium BC in the Dnieper valley (Mathieson et al. 2018).
The main problem is that this sample has more than one inconsistent, anachronistic data compared to its reported precise radiocarbon date ca. 4045–3974 calBCE (5215±20BP, PSUAMS-2832). I summarized them on Twitter:
- First known R1a-M417 sample, with subclade R1a-Y26 (Y2-), with formation date and TMRCA ca. 2750 BC (CI 95% ca. 3750–1950 BC), and proper of much later Steppe_MLBA bottlenecks. The closest available sample would be the Poltavka outlier of hg. R1a-Z94 (ca. 2700 BC), from a mixed cemetery that could belong to a later (likely Abashevo) layer; the closest related subclade is probably found in sample I12450 of Butkara_IA (ca. 800 BC).
- Ancestry and PCA cluster like Steppe_MLBA (see PCA below), different from neighbouring Sredni Stog samples of the roughly coetaneous Dereivka site (ca. 3600-3400 BC), and from a later Yamnaya sample from Dereivka (ca. 2800 BC), even more shifted toward WHG-related ancestry.
- Allele for lactase persistence (I3910-T), found only much later among Bell Beakers, and still later in Sintashta and Steppe_MLBA samples. This suggests a strong selection in northern Europe and South Asia stemming from steppe-related (and not forest-steppe-related) peoples, postdating the age of massive Indo-European migrations.
- Abashevo-like individuals (whose best proxy to date should be Poltavka_outlier I0432) potentially admixed with Poltavka-like herders; or
- Potapovka-like individuals potentially admixed with Catacomb-like peoples (whose best proxy until recently were probably Yamnaya_Kalmykia*).
- Yamnaya replaced Europeans, but admixed heavily as they spread to Asia
- Corded Ware culture origins: The Final Frontier
- Volga Basin R1b-rich Proto-Indo-Europeans of (Pre-)Yamnaya ancestry
- Yamnaya ancestry: mapping the Proto-Indo-European expansions
- Sredni Stog, Proto-Corded Ware, and their “steppe admixture”
- Aquitanians and Iberians of haplogroup R1b are exactly like Indo-Iranians and Balto-Slavs of haplogroup R1a
- Consequences of Damgaard et al. 2018 (II): The late Khvalynsk migration waves with R1b-L23 lineages
- The origin of social complexity in the development of the Sintashta culture
- The genetic and cultural barrier of the Pontic-Caspian steppe – forest-steppe ecotone
- Steppe and Caucasus Eneolithic: the new keystones of the EHG-CHG-ANE ancestry in steppe groups
- On the origin of haplogroup R1b-L51 in late Repin / early Yamna settlers
- On the origin and spread of haplogroup R1a-Z645 from eastern Europe
- Kurgan origins and expansion with Khvalynsk-Novodanilovka chieftains
- About Scepters, Horses, and War: on Khvalynsk migrants in the Caucasus and the Danube
- North Pontic steppe Eneolithic cultures, and an alternative Indo-Slavonic model
NOTE. The formation date of upper clade R1a-Z93 is estimated ca. 3000 BC, with a CI 95% ca. 3550–2550 BC, suggesting that the actual TMRCA range for the subclade has most likely a lower maximum formation date than estimated with the available samples under Y3.
Examining lactase persistence in the South and Central Asian data, shows that the allele arrives in South Asia with steppe pastoralists, (data from https://t.co/HPE0TOgd5a …) pic.twitter.com/kG66oRg23k
— Vagheesh Narasimhan (@vagheesh) April 18, 2018
Hajji Firuz Chalcolithic outlier
My impression is that the Hajji_Firuz Chalcolithic outlier, initially dated ca. 5900-5500 BC, had much less reason to be questioned than this sample, since Pre-Yamnaya ancestry was (and apparently is still) believed by members of the Reich Lab to have come from south of the Caucasus, and to have arrived around that time or earlier to the North Caspian steppe, i.e. before the 5th millennium BC.
The formation date of its initially reported haplogroup, R1b-Z2103, is ca. 4100 BC (CI 95% 4800-3500 BC), which seems also roughly compatible with that date and site – at least as compatible as R1a-Y3(xY2) is for ca. 4000 BC -, so it could have been interpreted as a migrant from the South Caspian region, potentially related to Proto-Anatolians, especially before the description of the Caucasus genetic barrier in Wang et al (2018). For some reason, though, the Hajji_Firuz sample was questioned, but this one didn’t even merited an interrogation mark.
There was already a similar situation with two samples (RISE568 and RISE569) initially reported as belonging to Czech Corded Ware groups, that turned out to be Early Slavs ca. 3,000 years younger, in turn more closely related to Bell Beaker-derived cultures of Central-East Europe. It seems little has changed since that case.
All in all, my guess is that genomic data of I6561 would have been a priori more compatible with a later period, during the expansion of East Corded Ware groups: at least Middle Dnieper culture, potentially Multi-Cordoned Ware culture, but most likely a Srubnaya-related one, given the most likely SNP mutation and TMRCA date, and the haplogroup variability found in the few samples available from that culture.
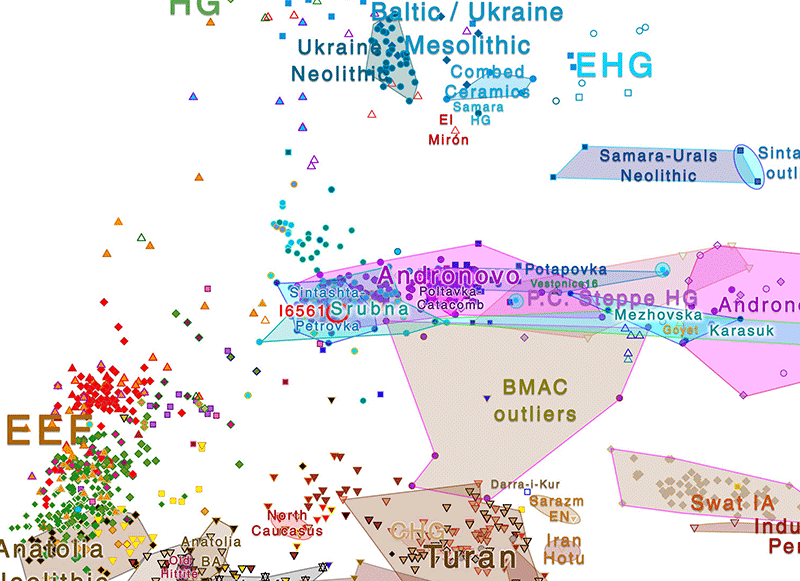
Compatibility checks
I tried to start a thread on the possibility that the radiocarbon date was wrong, and IF it were, how likely it would be that formal stats could actually show this, or how could we automatically prevent ancestry magic fiascos.
In other words: if this guy were a Srubnaya-related individual actually dated e.g. ca. 1700 BC, and someone would try to ‘prove’ – based on the current open source tools alone – that he was the ancestor of expanding peoples of the 4th and 3rd millennium BC (i.e. Balkan outliers, Yamnaya, Corded Ware, you name it), could these results be formally challenged?
I was hoping for some original brainstorming where people would propose crazy, essentially impossible to understand statistical models, say plotting dozens of well-studied mutations of different geographically related ancient samples with their reported dates, to visually highlight samples that don’t exactly fit with such a feature-based time series analysis; I mean, the kind of theoretical models I wouldn’t even be able to follow after the first two tweets or so. I didn’t receive an answer like that, but still:
Is it possible, with the currently available tools in population genomics, to tell whether this sample is ancestral to or derived from other Steppe-related populations, before the date is tested again? And how certain would these tests be…?
— Carlos Quiles (@cquilesc) September 14, 2019
@mathiesoniain may have more information, but it does seem to have a direct 14C date. In general, I'd be conservative about drawing strong inferences from a single outlier individual.
— Iosif Lazaridis (@iosif_lazaridis) September 14, 2019
The question in this case is more theoretical, though. IF this case were wrongly 14C-dated, would it be possible to detect this e.g. by assessing the number of mutations (like LP and the subclade) and the type of ancestry that shouldn't be there? More or less as w/ I2327
— Carlos Quiles (@cquilesc) September 14, 2019
I don't know how you would do it formally, but certainly if the ancestry is an outlier, it probably makes it more likely that the date is wrong.
— Iain Mathieson (@mathiesoniain) September 14, 2019
But, as far as LP is concerned, someone has to be the earliest, so I don't know that a single locus is strong evidence. Genome-wide ancestry a different story.
— Iain Mathieson (@mathiesoniain) September 14, 2019
Yfull dates are likely underestimated. I generally just multiply by 1.25. So, the subclade for this sample doesn't seem out of place. My opinion is that Sintashta and Andronovo don't come from CWC due to dated splits, etc., but formed in Eastern Europe.
— populationgenomicsblog (@populationgeno1) September 15, 2019
I have nothing to add to these answers, because I agree that all contradictory data are circumstancial.
#EDIT (Oct 14 2019) Iain Mathieson analysed ancient DNA from public data and wrote a post on the subject:
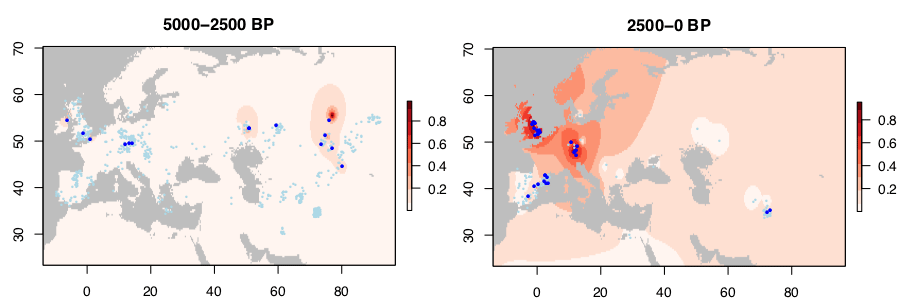
The current absolute lack of this kind of validity checks for ancestry models is disappointing, though, and leaves the so-called outliers in a dangerous limbo between “potentially very interesting samples” and “potentially wrongly dated samples”. Radiocarbon date is thus – together with compatibility of population source in terms of archaeological cultures and their potential relationship – a necessary variable to take into account in any statistical design: an error in one of these variables means a catastrophic error in the whole model.
Formal stats
For example, in these qpAdm models, I assumed Srubnaya, Ukraine_Eneolithic_outlier, and Bulgaria_MLBA samples were roughly coetaneous and potentially related to the Srubnaya-Sabatinovka–Noua cultural horizon, hence stemming from a source close to:
*To avoid adding more potential errors by merging different datasets, I have used only proxy samples available in the Reich Lab’s curated dataset of published ancient DNA.
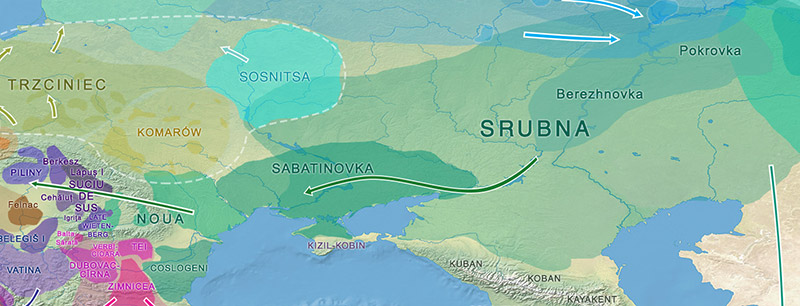
Apart from the lack of more models for comparison (I’m not going to dedicate more time to this), the results can’t be interpreted without a proper sampling and context, either, because (1) Poltavka_o may actually be from a much later group closely related to Srubnaya; (2) Bulgaria_MLBA is only one sample; and (3) there are only two samples from Potapovka; so the models here presented are basically useless, as many similar models that have been tested looking just for a formal “best fit”.
So feel free to chime in and contribute with ideas as to how to detect in the future whether a sample is ancestral to or derived from others. I will post here informative answers from Twitter, too, if there are any. I don’t think a discussion about the potentially wrong date in this specific sample is very useful, because this seems impossible to prove or disprove at this point. Just what tools or data would you use to at least try and assess whether samples are compatible with its reported date or not – preferably in some kind of automated sieve that takes dozens or hundreds of samples into account.
On the bright side, there is so much more than formal stats to arrive to relevant inferences about prehistoric populations, their movements and languages. That’s why I6561 didn’t matter for the conclusion by Anthony (2019) that it was the R1b-rich Eneolithic Don-Volga-Caucasus region the most likely Indo-Anatolian and Late Proto-Indo-European homeland, due to the creation of a wide Eneolithic mating network with extended exogamy practices, where Y-chromosome bottlenecks seem to be one of the main genomic data to take into account from the Neolithic to the Middle Bronze Age.
And that is the same reason why it doesn’t matter that much for the Proto-Indo-European or Uralic question for me, either.