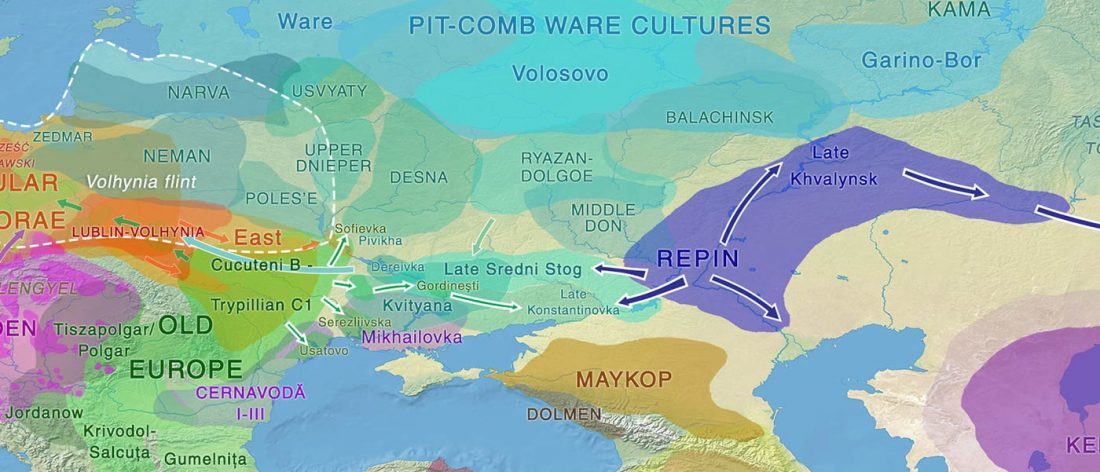
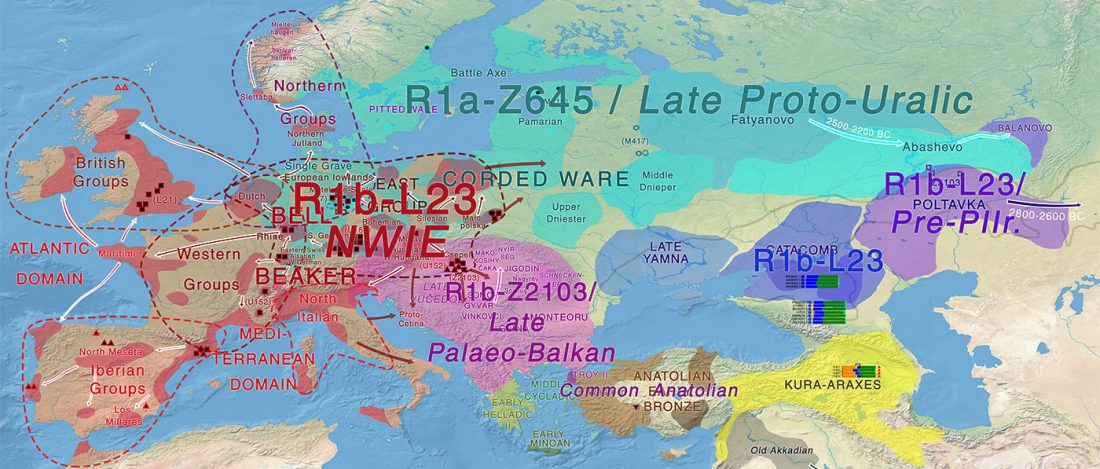
Over the past week or so, since the publication of new Corded Ware samples in Narasimhan, Patterson et al. (2019) and after finding out that the R1a-M417 star-like phylogeny may have started ca. 3000 BC, I have been ruminating the relevance of contradictory data about the Ukraine_Eneolithic_o sample from Alexandria, its potential wrong radiocarbon date, and its implications for the Indo-European question.
How many other similar ‘controversial’ samples are there which we haven’t even considered? And what mechanisms are in place to control that the case of Hajji_Firuz_CA I2327 is not repeated?
Ukraine Eneolithic outlier I6561
It was not … Read the rest “On the Ukraine Eneolithic outlier I6561 from Alexandria”