Yamnaya ancestry: mapping the Proto-Indo-European expansions
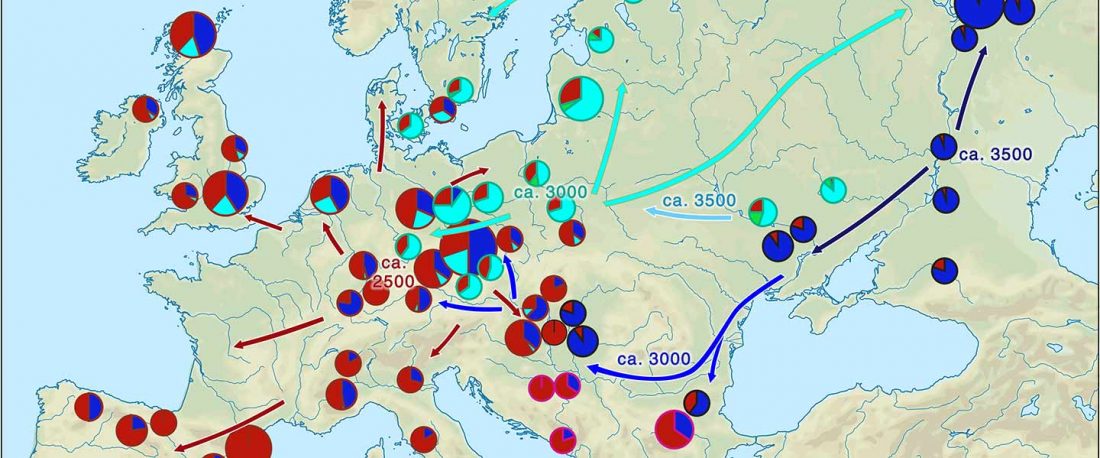
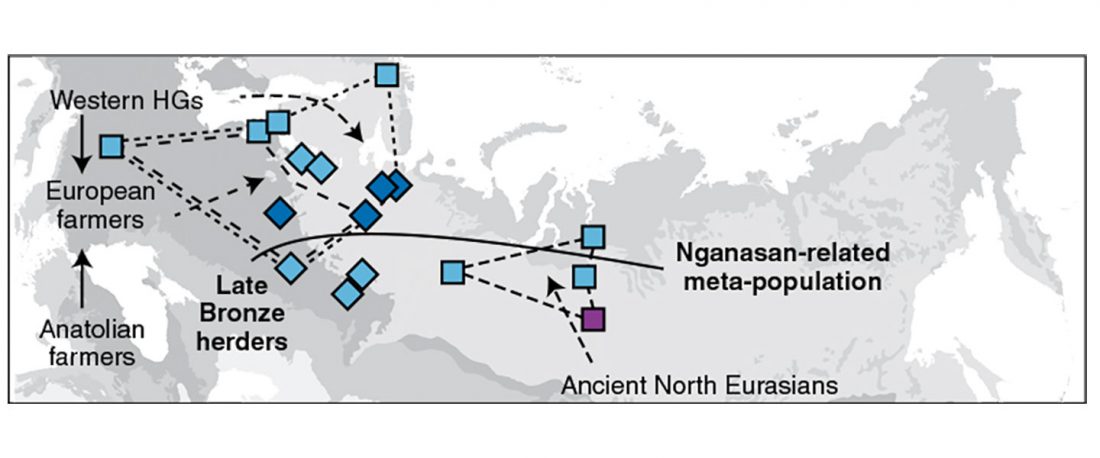
The latest papers from Ning et al. Cell (2019) and Anthony JIES (2019) have offered some interesting new data, supporting once more what could be inferred since 2015, and what was evident in population genomics since 2017: that Proto-Indo-Europeans expanded under R1b bottlenecks, and that the so-called “Steppe ancestry” referred to two different components, one – Yamnaya or Steppe_EMBA ancestry – expanding with Proto-Indo-Europeans, and the other one – Corded Ware or Steppe_MLBA ancestry – expanding with Uralic speakers.
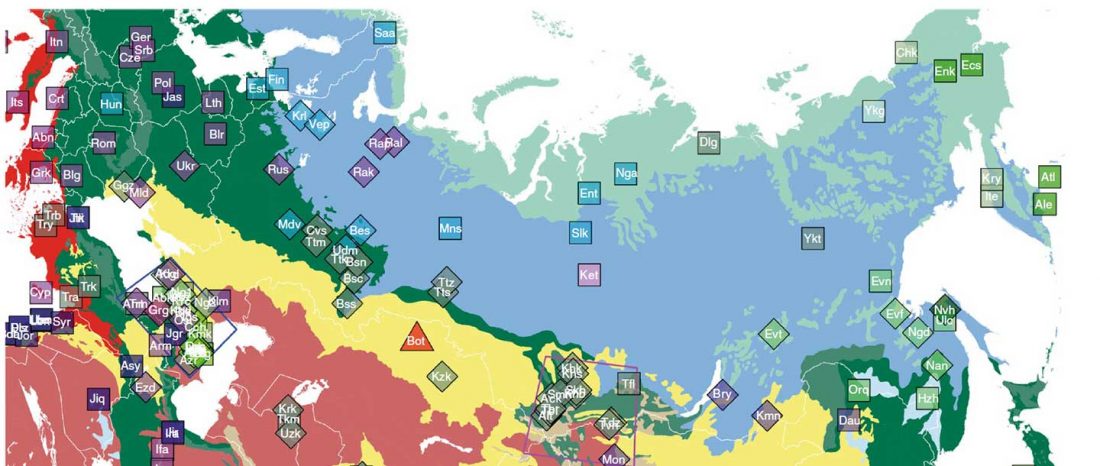
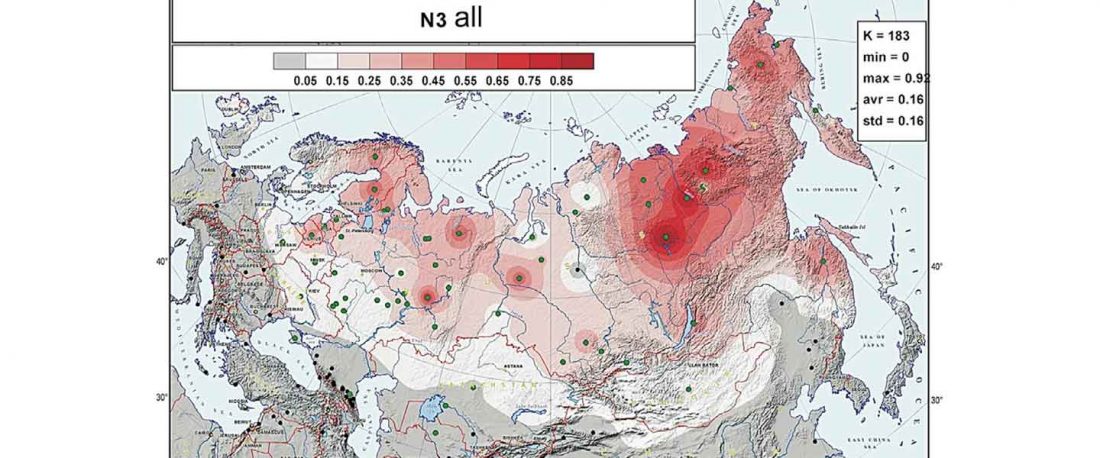
The following maps are based on formal stats published in the papers and supplementary materials from 2015 until today, mainly on … Read the rest “Yamnaya ancestry: mapping the Proto-Indo-European expansions”