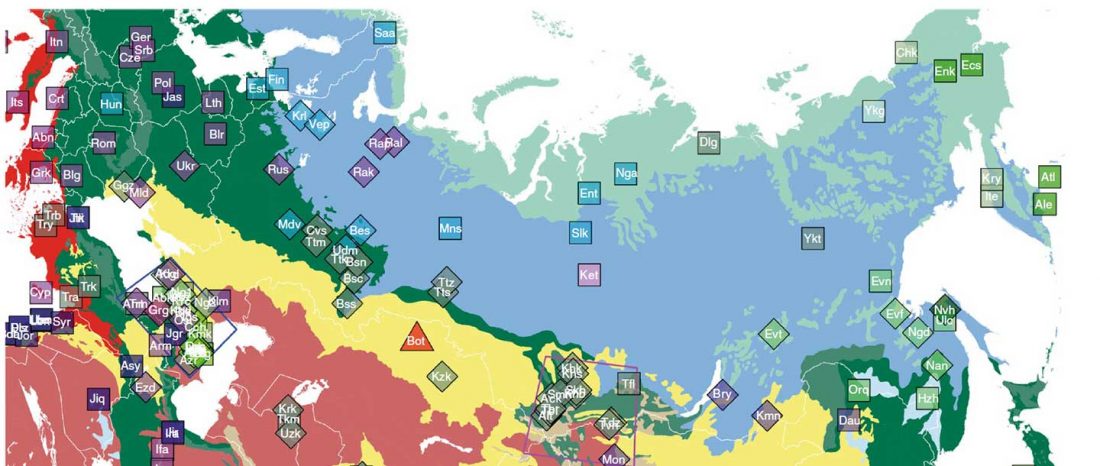
The preprint by Jeong et al. (2018) has been published: The genetic history of admixture across inner Eurasia Nature Ecol. Evol. (2019).
Interesting excerpts, referring mainly to Uralic peoples (emphasis mine):
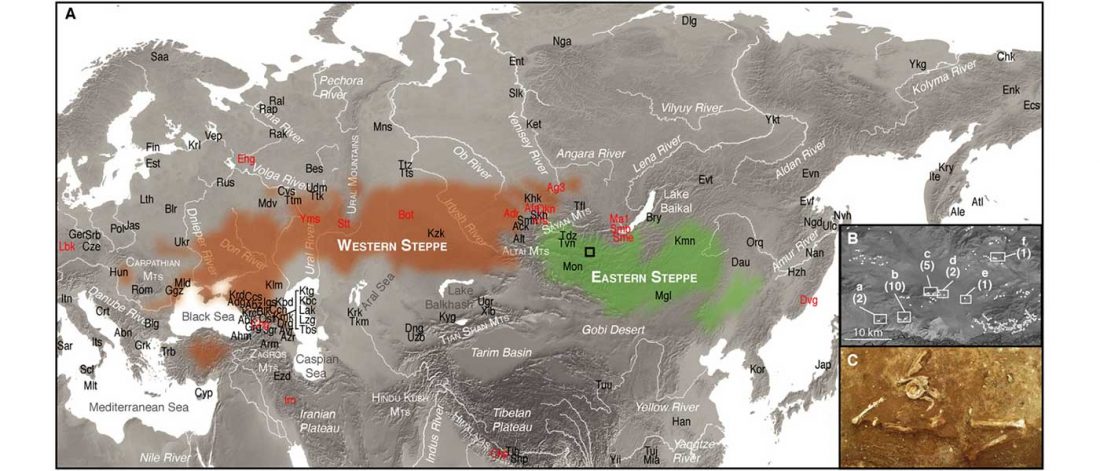
… Read the rest “Uralic speakers formed clines of Corded Ware ancestry with WHG:ANE populations”A model-based clustering analysis using ADMIXTURE shows a similar pattern (Fig. 2b and Supplementary Fig. 3). Overall, the proportions of ancestry components associated with Eastern or Western Eurasians are well correlated with longitude in inner Eurasians (Fig. 3). Notable outliers include known historical migrants such as Kalmyks, Nogais and Dungans. The Uralic- and Yeniseian-speaking populations, as well as Russians from multiple locations, derive most of their Eastern Eurasian ancestry