New preprint The population history of northeastern Siberia since the Pleistocene, by Sikora et al. bioRxiv (2018).
Interesting excerpts (emphasis mine; most internal references removed):
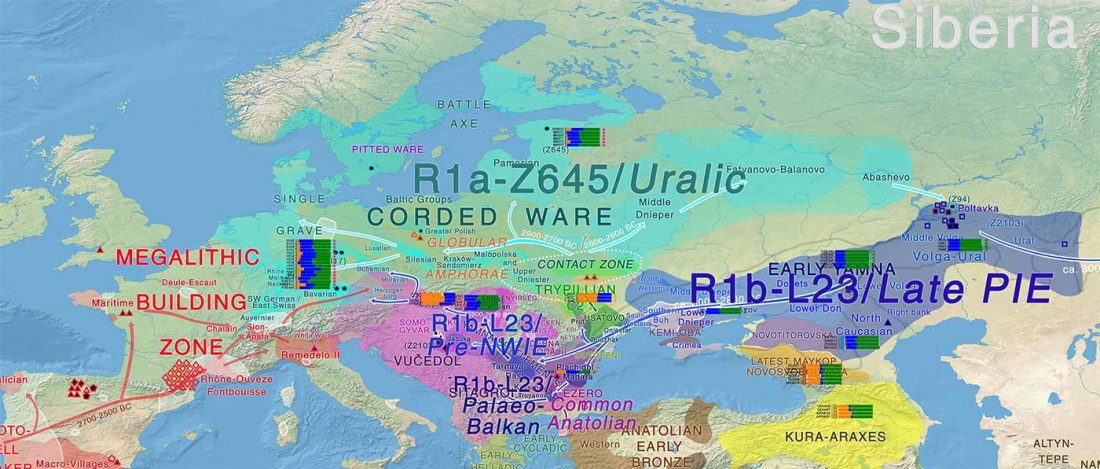
ANE ancestry
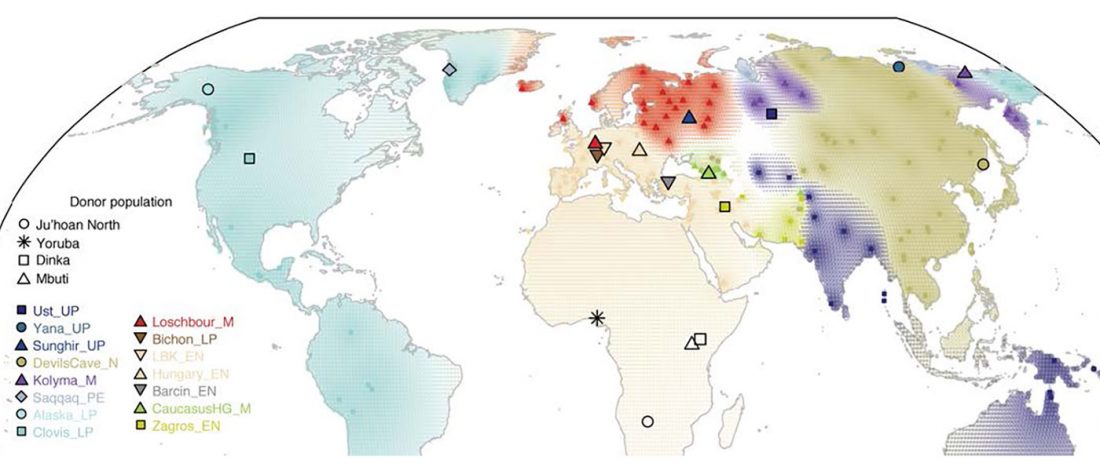
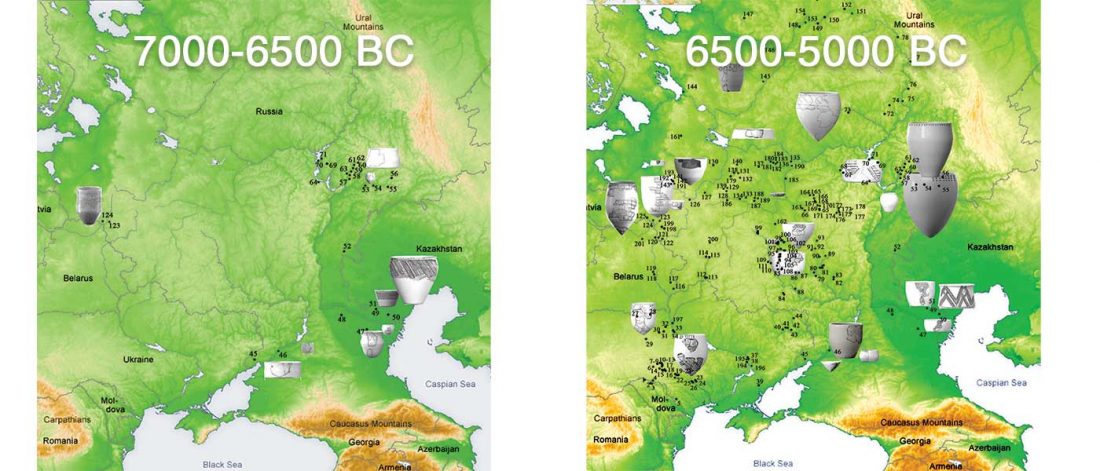
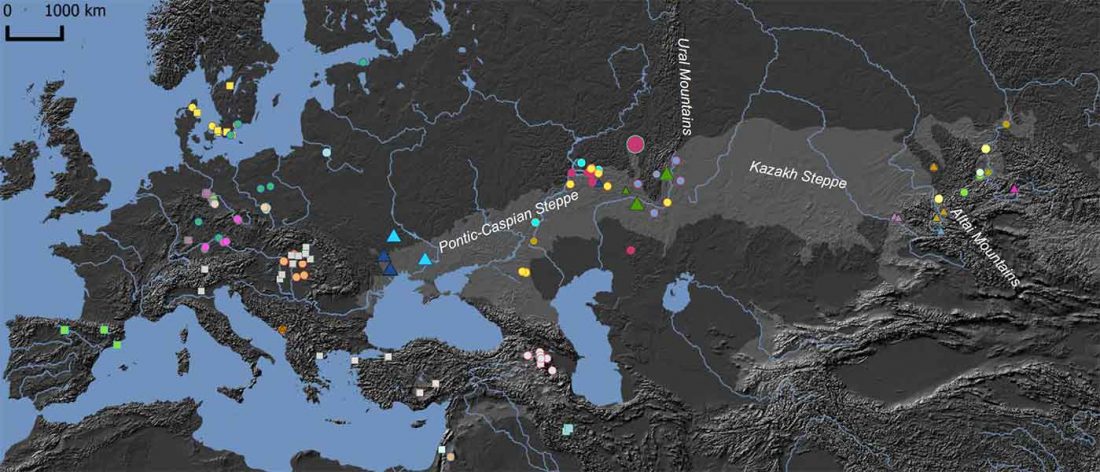
The earliest, most secure archaeological evidence of human occupation of the region comes from the artefact-rich, high-latitude (~70° N) Yana RHS site dated to ~31.6 kya (…)
… Read the rest “Waves of Palaeolithic ANE ancestry driven by P subclades; new CWC-like Finnish Iron Age”The Yana RHS human remains represent the earliest direct evidence of human presence in northeastern Siberia, a population we refer to as “Ancient North Siberians” (ANS). Both Yana RHS individuals were unrelated males, and belong to mitochondrial haplogroup U, predominant among ancient West Eurasian hunter-gatherers,