New preprint Early Medieval Genetic Data from Ural Region Evaluated in the Light of Archaeological Evidence of Ancient Hungarians, by Csaky et al. bioRxiv (2020).
Interesting excerpts (emphasis mine):
Based on linguistic evidences, the Hungarian language, belonging to the Ugric branch of the Uralic language family, was developed at the eastern side of Ural Mountains between 1000-500 BC. According to the written and linguistic sources and archaeological arguments, after the 6th century AD, part of the predecessors of Hungarians moved to the Western Urals (Cis-Ural region) from their ancient homeland. Around the first third of 9th century AD a part of this Cis-Uralic population crossed the Volga-river and settled near to the Khazarian Khaganate in the Dnieper-Dniester region. Early Hungarians lived in Eastern Europe (forming the so-called Subbotsy archaeological horizon) until the conquest of the Carpathian Basin that took place in 895 AD. The material traits of 10th century AD Carpathian Basin was rapidly transformed after the conquest, its maintained cultural connections with East-European regions have numerous doubtless archaeological evidence
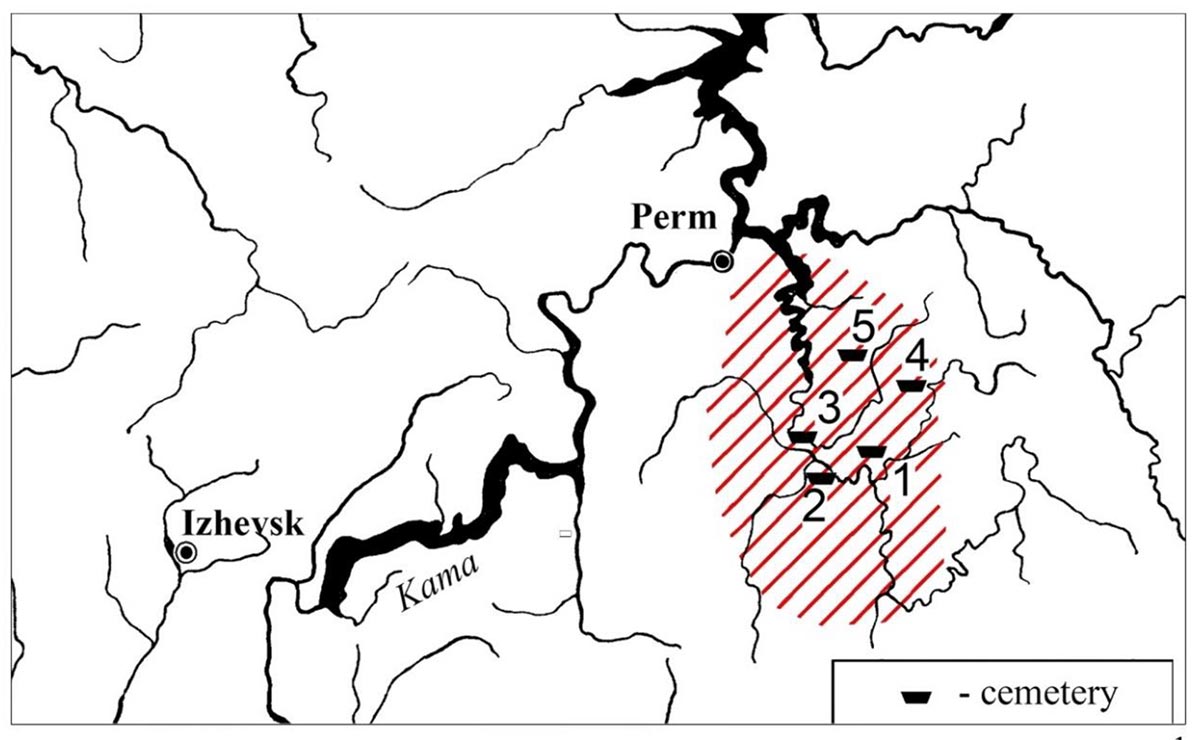
Nevolino and Kushnarenkovo culture
As the archaeological and historical theories are slightly diverse, we aimed to cover a wide range of early medieval archaeological cultures located in the middle course of the Kama river in the west side of the Ural Mountains (Cis-Ural region). Scholars connect the termination of the Nevolino Culture in 8-9th centuries AD to the westward migration of ancestors of Hungarians, hence the sampling was carried out in all three phases of this culture: Brody (3rd-4th centuries), Bartym (5-6th centuries) and Sukhoy Log (7-8th centuries). Furthermore, we investigated the Bayanovo cemetery (9-10th centuries AD), which represents the southern variant of Lomovatovo culture that shows close cultural connection to its southern neighbour Nevolino culture. The sampling of the richly furnished graves of Bayanovo was limited by the poor preservation of bone samples.
The sampled Uyelgi cemetery from Trans-Ural region presented the greatest similarity to the archaeological traits of the tenth-century Carpathian Basin. This cemetery of the late Kushnarenkovo culture was used between the end of 8th century to 11th century.
Corded Ware ancestry
We performed genomic PCA of five Uyelgi samples consisting of 10,828 nuclear genomic SNPs on average gained from 3000 SNP capture and shallow shotgun sequencing data (from 598,094 called SNPs). The five samples are plotted together on the genomic PCA and they also appear close to the modern Bashkir and Siberian Tatar individuals as well as to the Altaian Bronze Age Okunevo population, to a hunter-gatherer individual from Tyumen region and Iron Age Central Sakas from Kazakhstan and in line with the uniparental makeup.
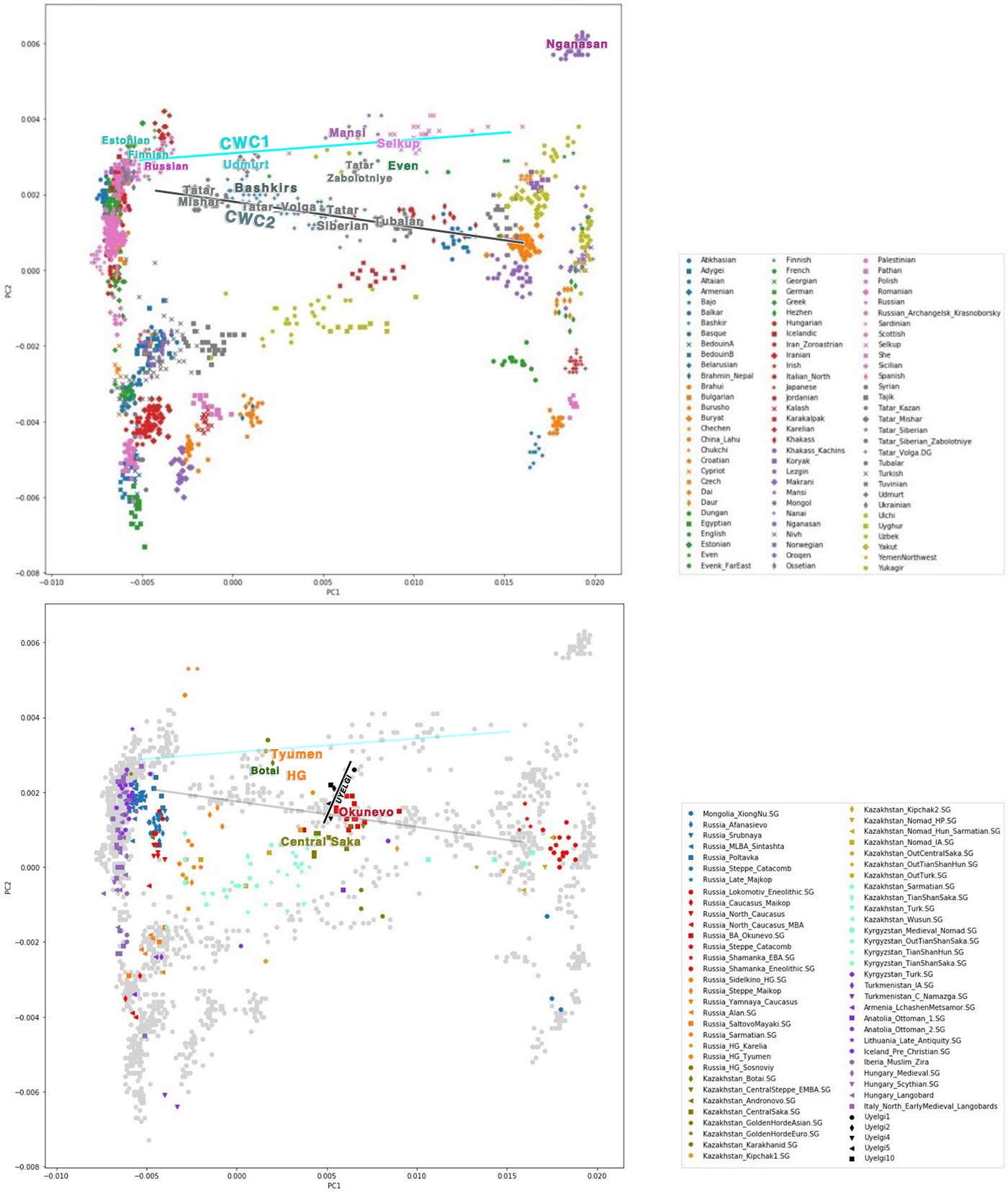
Iranian- vs. Turkic-related Admixture
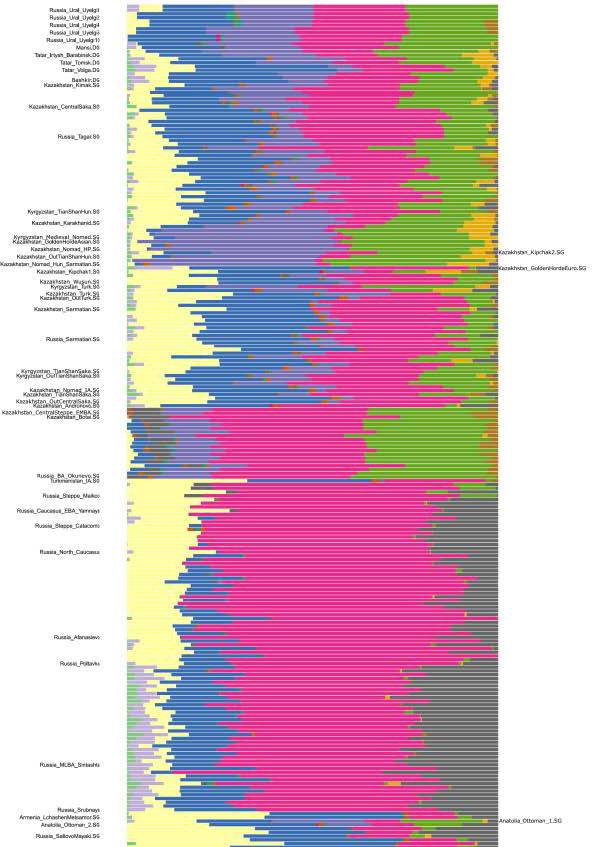
We can describe the affinities of ancestry components of Uyelgi samples through those modern populations in that the five samples’ ancestry components are maximized: Uyelgi population has modern European-related components, a modern North-Siberian-related, an East-Asian-related component and a component that is maximised in hunter gatherer samples from Tyumen and Afontova Gora3.
The five Uyelgi samples with an average of 22,540 SNPs show the most similar ancestry cluster proportions to present-day Mansis and Irtysh-Barabinsk Tatars. The structure of ancestry components is different from Uyelgi in case of the Okunevo that shows more North-Siberian and East-Asian related ancestry. Closest ancient proxies in the current published databases are the Iron Age Central Sakas and early medieval Kimak from present-day North Kazakhstan. The Central Sakas (Inner Asian Scythians) were modelled as a two-way mixture of Late Bronze Age pastoralists (56%) and southern Siberian hunter-gatherers (44%) by Damgaard et al., which scenario is plausible for the Uyelgi population as well.
mtDNA haplogroup
The new dataset consists of the mixture of nine macrohaplogroups (A, C, D, H, T, U, N, R, Z) (Fig. 3a). Haplogroups of presumably west Eurasian origin are represented by U (U2e1, U3a1, U4a1d, U4b1a1a1, U4d2, U5a1a1, U5b2a1a1, N=12), H (H1b2, H3b, H40b, N=9), N (N1a1a1a1a, N=5) and T (T1a1, T1a2, T2b4h, N=5), although phylogeographic analyses show eastern origin for some of them, see Table1 and Supplementary Figs. S4a-s. Eastern Eurasian lineages are represented by A (A+152+16362, A12a, N=4), C (C4a1a6, C4a2a1, N=6), D (D4j, D4j2, N=2), along with R11b1b and Z1a1a by one individual each.
Y-chromosome haplogroup
In contrast to the mitochondrial lineages, the Y-chromosomal gene pool based on STR and/or SNP data show homogenous composition in our dataset: 83.3% is N-M46, 5.5% G2a (G176 L1266), 5.5% J2 and 5.5% is R1b of the typed male individuals (Supplementary Table S2). 13 male samples out of 19 from Uyelgi cemetery carry Y-haplogroup N with various DNA preservation-dependent subhaplogroup classifications, while in the Cis-Ural we detected three N-M46 Y-haplogroups (samples from Brody, Bartym and Bayanovo cemeteries). The overall poor preservation of further Cis-Uralic samples from Sukhoy Log and Bartym disabled further Y-chromosome based analyses.
Seven samples of Uyelgi site most probably belong to N-Y24365 (also known as N-B545 and N1a1a1a1a2a1c2 in ISOGG 14.255) under N-Z1936, a specific subclade that can be found almost exclusively in todays’ Tatarstan, Bashkortostan and Hungary (ISOGG, Yfull).
Median Joining (MJ) network analysis is 346 performed using 238 N-M46 Y-haplotypes including seven samples from Uyelgi detected with 17 STR loci as well as 335 N-M46 Y-haplotypes with 12 STR loci. Based on MJ of 17 Y-STR loci, certain samples show identical or one-step neighbour profiles to Bashkirs, Khantys, Hungarians, Tatars from Volga-Ural region and a Central Russian sample. The MJ based on 12 Y-STR data show one-step neighbour connection of Uyelgi with two Hungarian conquerors from Bodrogszerdahely-Bálványhegy and Karos-Eperjesszög.
In the Cis-Ural sample set the DNA preservation was insufficient for proper paternal lineage analyses, the only obtained N-M46 Y-haplotype of Bay2 sample and the R1b haplotype of Bartym3 do not have direct matches in the worldwide YHRD database, however, we found four one-step-neighbours of Bay2 from Sverdlovsk Oblast (Ural region) and Lithuania.
#EDIT: The reported Y-DNA and mtDNA of Cis- and Trans-Uralian samples from this study are already on the Ancient DNA Dataset, and on the ArcGIS Online Web App. Here is how these Y-DNA samples would complete the maps:
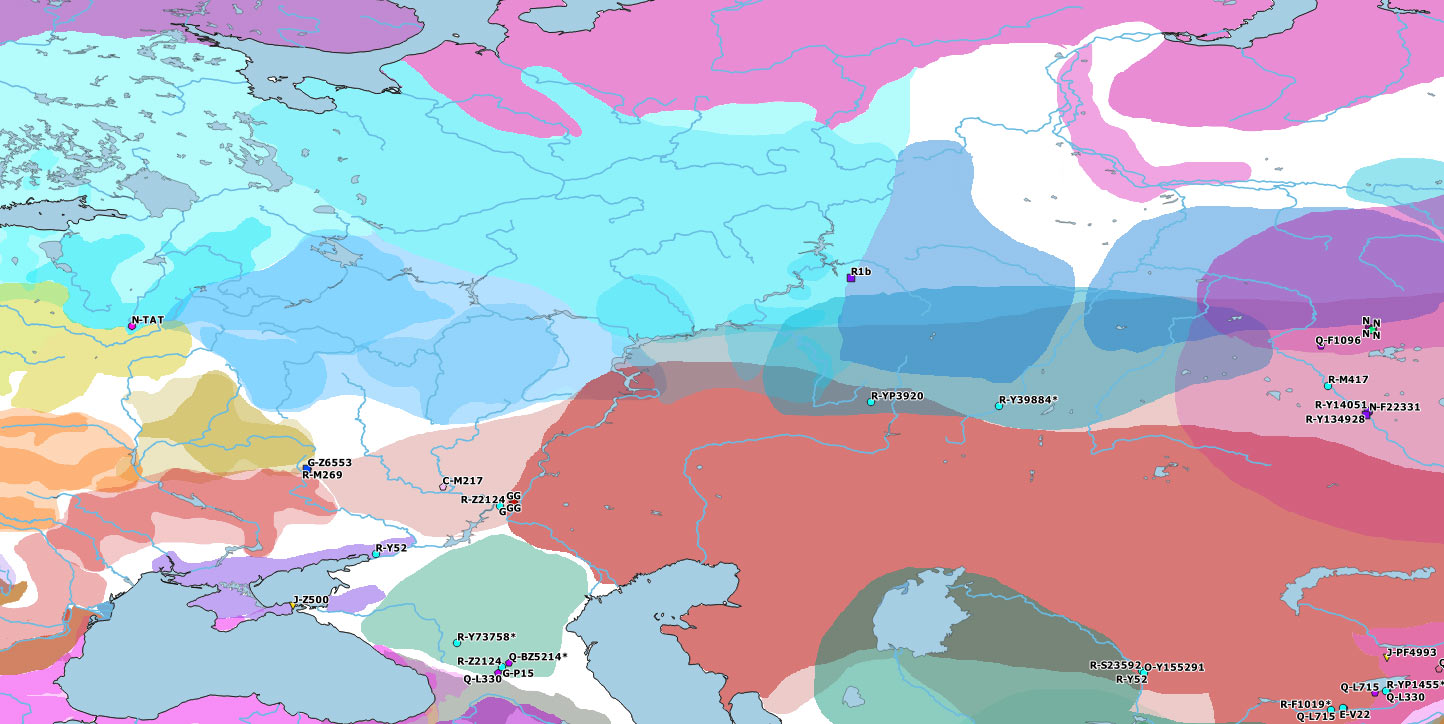
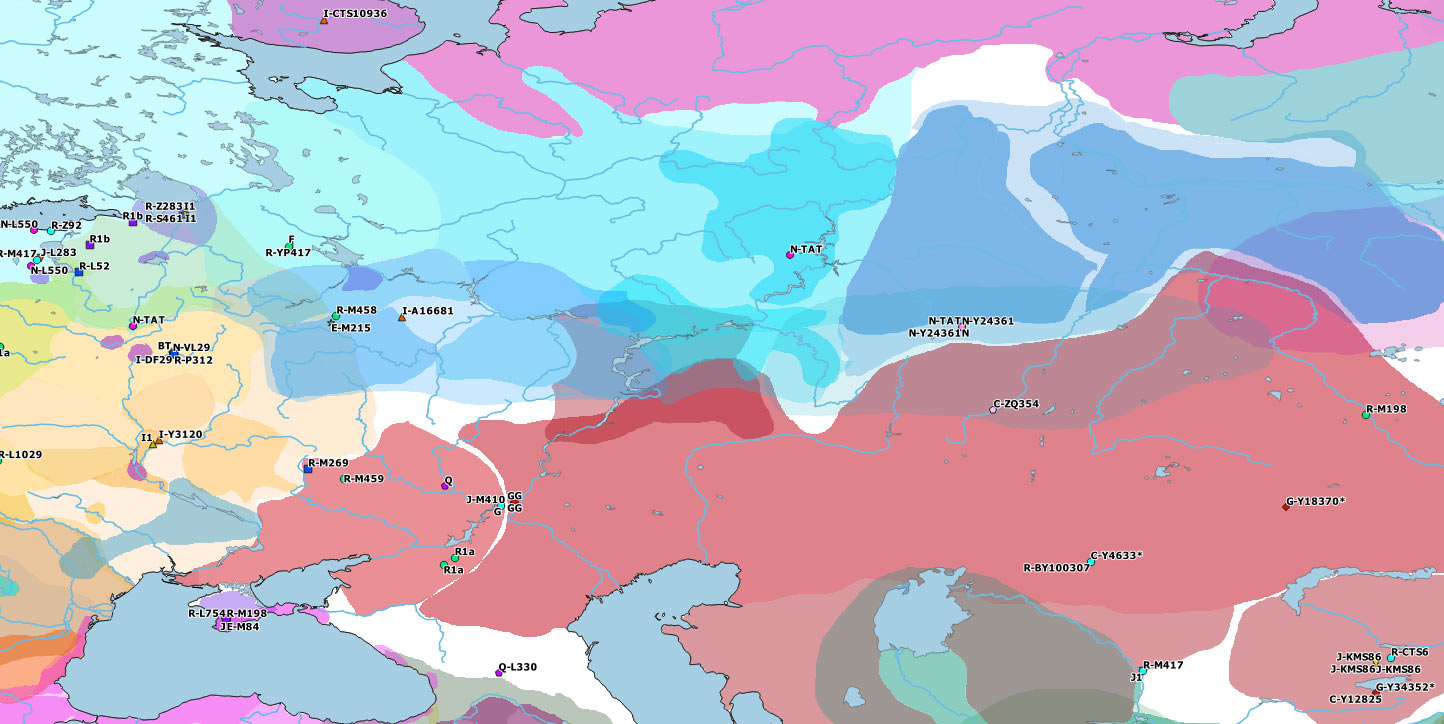
For more on the evolution of Ugric peoples, see: cultures of the Eurasian forest-steppes and more specifically Ugrians and Samoyeds
Related
- More Hungarian Conquerors of hg. N1c-Z1936, and the expansion of ‘Altaic-Uralic’ N1c
- Baltic Finns in the Bronze Age, of hg. R1a-Z283 and Corded Ware ancestry
- Uralic speakers formed clines of Corded Ware ancestry with WHG:ANE populations
- The cradle of Russians, an obvious Finno-Volgaic genetic hotspot
- Magyar tribes brought R1a-Z645, I2a-L621, and N1a-L392(xB197) lineages to the Carpathian Basin
- R1a-Z280 and R1a-Z93 shared by ancient Finno-Ugric populations; N1c-Tat expanded with Micro-Altaic
- The complex origin of Samoyedic-speaking populations
- Corded Ware—Uralic (IV): Hg R1a and N in Finno-Ugric and Samoyedic expansions
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- The traditional multilingualism of Siberian populations
- Corded Ware—Uralic (I): Differences and similarities with Yamna
- Haplogroup R1a and CWC ancestry predominate in Fennic, Ugric, and Samoyedic groups