Looking for differences among steppe cultures in Genomics is like looking for a needle in a haystack.
It means, after all, looking for differences among closely related cultures, such as between South-Western and North-Western Anatolian Neolithic cultures, or among Old European cultures (such as Vinča or Cucuteni–Trypillia), or between Iberian cultures after the arrival of steppe-related populations.
These differences between closely related regions, in all these cases and especially among steppe cultures, even when they are supported by Archaeology and anthropological models of migration (and compatible with linguistic models), are expected to be minimal.
Fortunately, we have phylogeography, which helps us point in the right direction when assessing potential migrations using genomic data.
User Tomenable recently pointed out a curious finding on Anthrogenica, from data available in Mathieson et al (2017): in ADMIXTURE results with K=12, a different ancestral component (in light green in the paper, see below) is traceable from the North Caspian steppe since the Neolithic. This is also partially distinguishable on K=10 and K=11, although not so clearly differentiating among later cultures.
NOTE: Read more on the controversy regarding the ideal number of ancestral populations, the absurd use of ADMIXTURE to solve language questions, and the meaning of cross-validation (CV) values
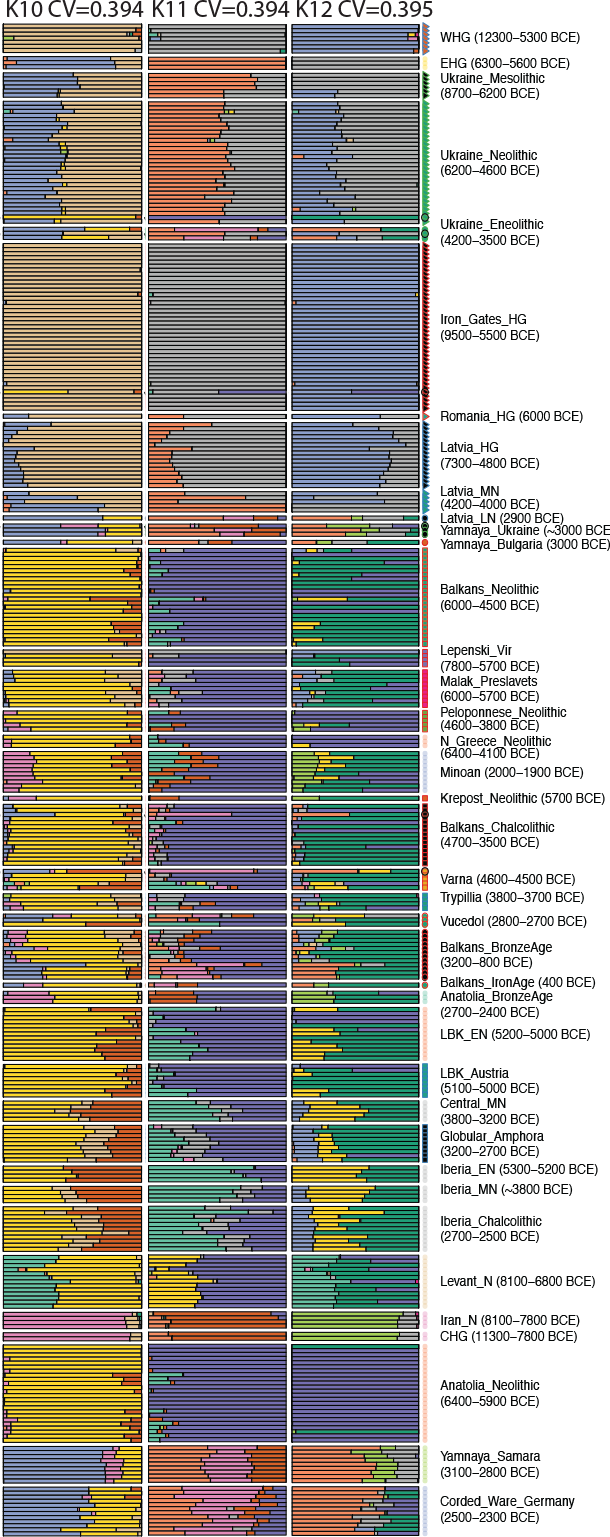
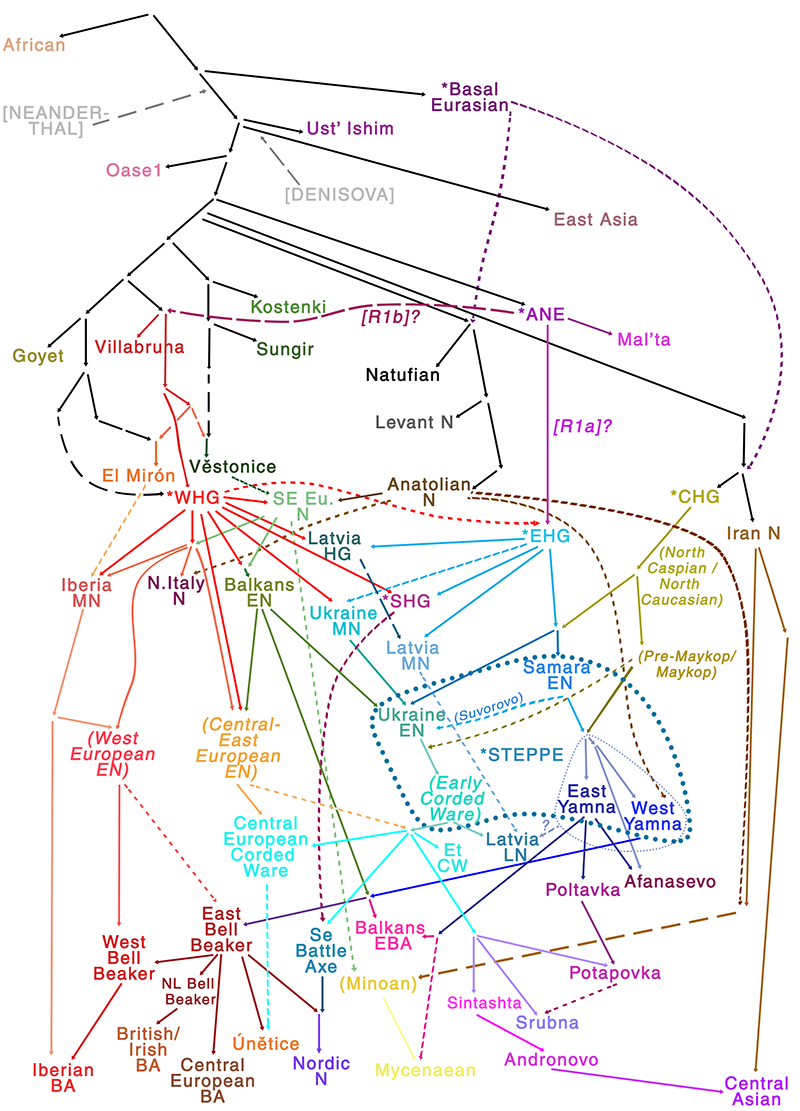
Explanations for this finding might include, as the user points out, a greater contribution of CHG ancestry in the eastern steppe cultures (Khvalynsk/Yamna) compared to the North Pontic steppe (Sredni Stog/Corded Ware), which is probably one of the main genomic differences among both cultures, as I pointed out in the Indo-European demic diffusion model (see accounts on the origins of Khvalynsk and Sredni Stog populations and on contacts between Yamna and the Caucasus, and see below also my sketch of Eurasian genomic history).
Interesting is also the appearance of similar ancestral components later in Vučedol – which probably received admixture from Yamna settlers (see admixture components in West Yamna samples and in the Yamna settler from Bulgaria) – , and later still in the Balkans.
On the other hand, previous ancestral components in outliers from the Balkans seem to be more similar to Sredni Stog samples, giving still more strength to the hypothesis that this common (“steppe”) component expanded westward within the Pontic-Caspian steppe with the spread of Suvorovo-Novodanilovka chiefs.
Problems with this interpretation include:
1) The scarce samples available, the different cultures included, and the CV values of the K populations selected in ADMIXTURE.
2) The lack of data for comparison with Bell Beaker peoples (from Olalde et al. 2017).
3) The sample classified as Latvia_LN/CWC has this component. I have already said before that, given the differences with all other Corded Ware samples, this quite early sample might be an outlier, with Khvalynsk/Yamna population connected directly to the ancestors of this individual, possibly through exogamy (as it is clear from my sketch below). Whether or not this is an outlier among CWC populations in the Baltic, only future samples can tell.
4) Three later individuals from Corded Ware in Germany have the component, in a minimal amount. I would bet – judging by their position in the graphic – that this might be explained through the Esperstedt family. These individuals might have in turn got the contribution directly from the oldest member, who shows what seems (in PCA) like a recent admixture from contemporary steppe cultures (such as the Catacomb culture).
NOTE: See my graphics with interesting members of the Espersted family marked: ADMIXTURE and PCA (outlier).
Again, needle in a haystack… And confirmation bias by me, indeed.
But interesting nonetheless.
EDIT (4 JAN 2017): A reader points out that the interpretation of Unsupervised ADMIXTURE should work backwards (i.e. different contributions into different modern populations), and not based solely on ancestral populations, which seems probably right. So again, confirmation bias (and potentially wrong direction fallacy) by me…
Related:
- Human ancestry solves language questions? New admixture citebait
- The new “Indo-European Corded Ware Theory” of David Anthony
- The renewed ‘Kurgan model’ of Kristian Kristiansen and the Danish school: “The Indo-European Corded Ware Theory”
- Correlation does not mean causation: the damage of the ‘Yamnaya ancestral component’, and the ‘Future America’ hypothesis
- Something is very wrong with models based on the so-called ‘steppe admixture’ – and archaeologists are catching up
- New Ukraine Eneolithic sample from late Sredni Stog, near homeland of the Corded Ware culture
- Germanic–Balto-Slavic and Satem (‘Indo-Slavonic’) dialect revisionism by amateur geneticists, or why R1a lineages *must* have spoken Proto-Indo-European