New paper (behind paywall), The genomic history of the Iberian Peninsula over the past 8000 years, by Olalde et al. Science (2019).
NOTE. Access to article from Reich Lab: main paper and supplementary materials.
Abstract:
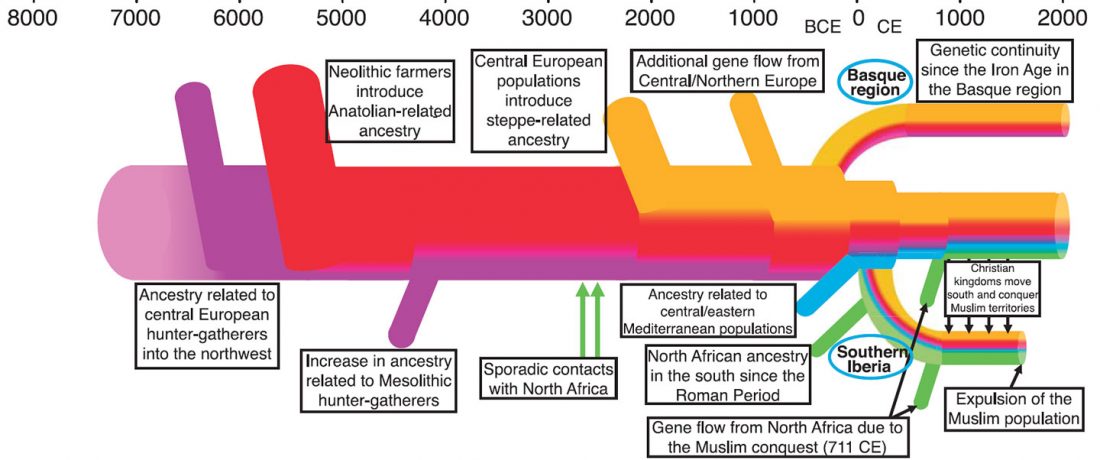
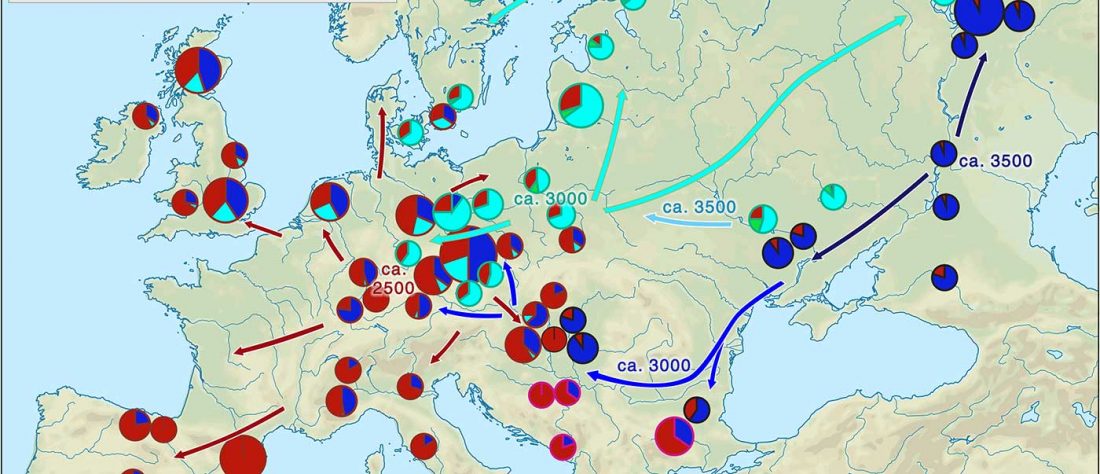
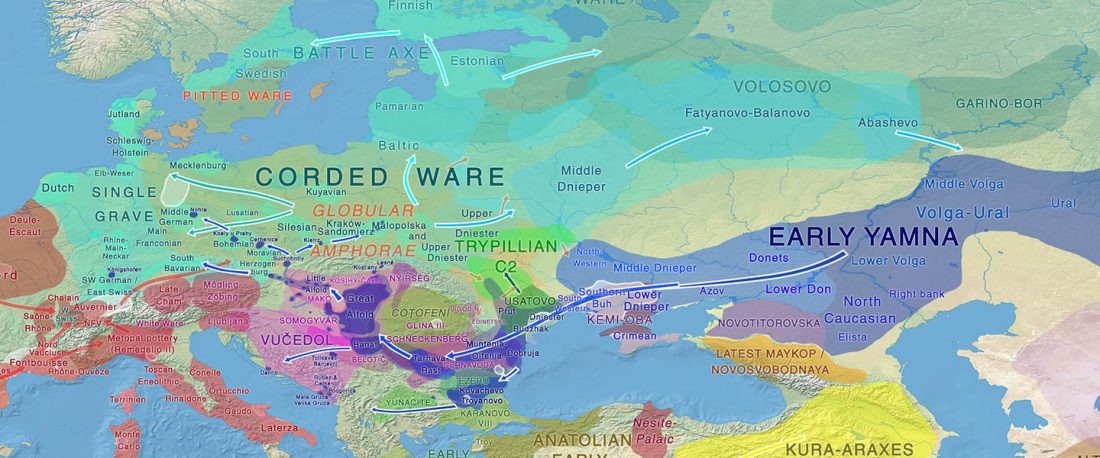
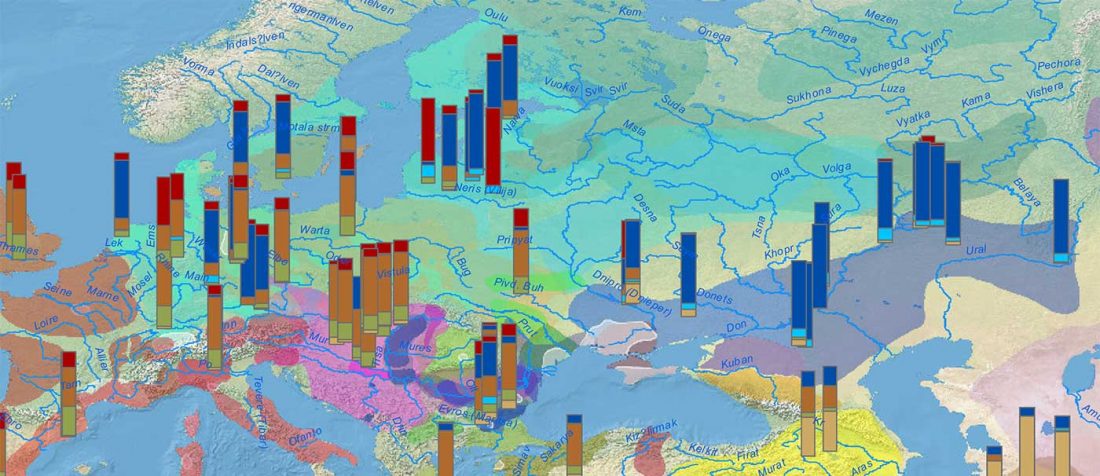
… Read the rest “Iberia: East Bell Beakers spread Indo-European languages; Celts expanded later”We assembled genome-wide data from 271 ancient Iberians, of whom 176 are from the largely unsampled period after 2000 BCE, thereby providing a high-resolution time transect of the Iberian Peninsula. We document high genetic substructure between northwestern and southeastern hunter-gatherers before the spread of farming. We reveal sporadic contacts between Iberia and North Africa by ~2500 BCE and, by ~2000 BCE, the replacement