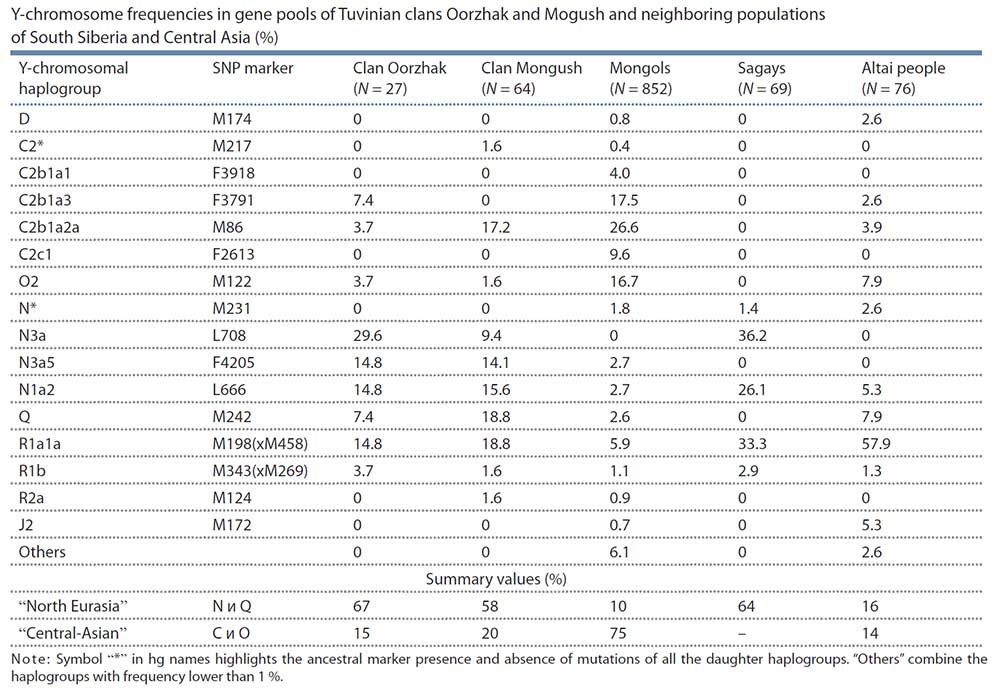
Open access Estimating the impact of the Mongol expansion upon the gene pool of Tuvans, by Balanovskaya et al., Vavilov Journal of genetics and breeding (2018), 22(5):611-619.
Abstract (emphasis mine):
With a view to trace the Mongol expansion in Tuvinian gene pool we studied two largest Tuvinian clans – those in which, according to data of humanities, one could expect the highest Central Asian ancestry, connected with the Mongol expansion. Thus, the results of Central Asian ancestry in these two clans component may be used as upper limit of the Mongol influence upon the Tuvinian gene pool in a whole. According to the data of 59 Y-chromosomal SNP markers, the haplogroup spectra in these Tuvinian tribal groups (Mongush, N = 64, and Oorzhak, N = 27) were similar. On average, two-thirds of their gene pools (63 %) are composed by North Eurasian haplogroups (N*, N1a2, N3a, Q) connected with autochtonous populations of modern area of Tuvans. The Central Asian haplogroups (C2, O2) composed less then fifth part (17 %) of gene pools of the clans studied. The opposite ratio was revealed in Mongols: there were 10 % North Eurasian haplogroups and 75 % Central Asian haplogroups in their gene pool. All the results derived – “genetic portraits”, the matrix of genetic distances, the dendrogram and the multidimensional scaling plot, which mirror the genetic connections between Tuvinian clans and populations of South Siberia and East Asia, demonstrated the prominent similarity of the Tuvinian gene pools with populations from and Khakassia and Altai. It could be therefore assumed that Tuvinian clans Mongush and Oorzhak originated from autochtonous people (supposedly, from the local Samoyed and Kets substrata). The minor component of Central Asian haplogroups in the gene pool of these clans allowed to suppose that Mongol expansion did not have a significant influence upon the Tuvinan gene pool at a whole.
Interesting excerpts:
Haplogroup C2 peaks in Central Asia (Wells et al., 2001; Zerial et al., 2003), though its variants are abundant in other peoples of Siberia and Far East. For instance, in one of Buryat clans, namely Ekhirids, hg C2 frequency is 88 % (Y-base); in Kazakhs from different regions of Kazakhstan, total occurrence of hg C2 variants averages between 17 and 81 % (Abilev et al., 2012; Zhabagin et al., 2013, 2014, 2017), in populations of the Amur River (such as Nanais, Negidals, Nivkhs, Ulchs) – between 40 and 65 %, in Evenks – up to 68 % (Y-base), in Kyrgyz people of Pamir-Alay – up to 22 %, correspondingly; of all Turkic peoples of Altai, relatively high hg C2 frequency (16 %) is detected only in Telengits (Balanovskaya et al., 2014; Balaganskaya et al., 2011a, 2016). In Tuvinian clans under the study, hg C2 frequency is rather low – 19 % in Mongush and 11 % in Oorzhak, while in Mongols it makes up almost two thirds of the entire gene pool an comprises different genetic lines (subhaplogroups).
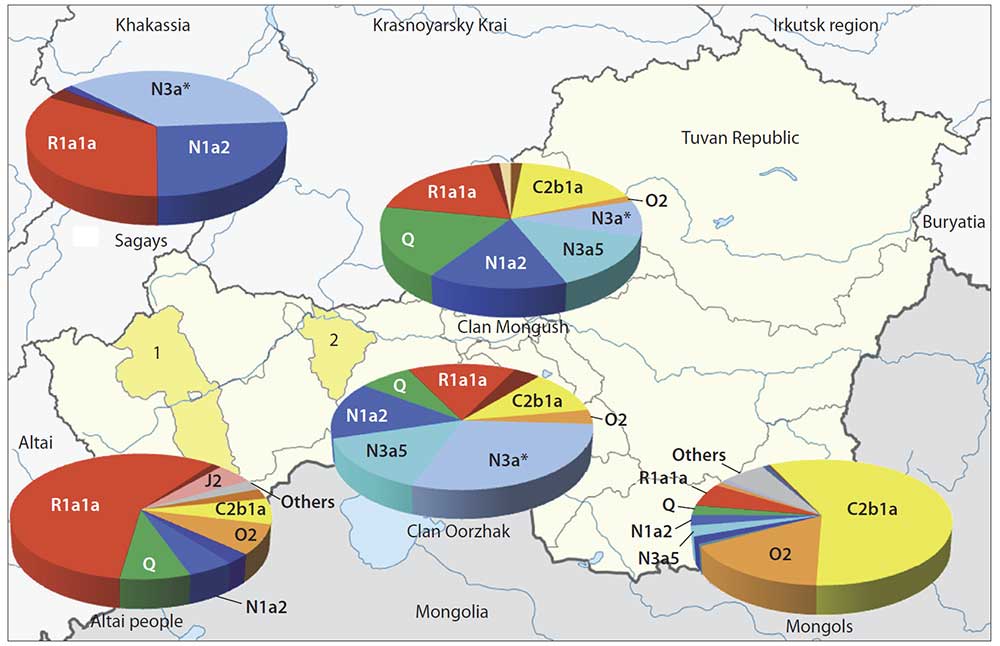
Haplogroup N is abundant all over North Eurasia from Scandinavia to Far East (Rootsi et al., 2007). The study on whole Y-chromosome sequencing conducted with participation of our group (Ilumäe et al., 2016) subdivided this haplogroup into several branches with their regional distribution. In gene pools of the Tuvans involved, hg N was represented by two sub-clades, namely N1a2 and N3a.
Sub-clade N1a2 peaks in populations of West Siberia (in Nganasans, frequency is 92 %) and South Siberia (in Khakas 34 %, in Tofalars 25 %) (Y-base). In Tuvans, N1a2 occurrence is nearly 16 % in Mongush and 15 % in Oorzhak clans, respectively, while in Mongols, the frequency is three times less (5 %). Hg N1a2 is supposed to display the impact of the Samoyedic component to the gene pool of Tuvinian clans (Kharkov et al., 2013).
Sub-clade N3a is major in the Oorzhak clan comprising almost half of the gene pool (45 %); it is represented by two sub-clades, namely N3a* and N3a5. The same sub-branches are specific to the Mongush clan as well, though with lower frequencies: N3a* – 9 % and N3a5 – 14 % (see Table). In Khori-Buryats from the Transbaikal region, a high frequency is observed – 82 % (Kharkov et al., 2014), while in Mongols, N3a5 occurs rather rarely (6 %). Hg N3a* was detected in populations of South Siberia only, and was widely spread in Khakas-Sagays and Shors (up to 40 %) (Ilumäe et al., 2016) (Y-base).
Within the pan-Eurasian haplogroup R1a1a, two large genetic lines (sub-haplogroups) are identified: “European” (marker M458) and “Asian” (marker Z93) the latter almost never occurring in Europe (Balanovsky, 2015) but abundant in South Siberia and northern Hindustan. In the Altai-Sayan region, high frequencies of the “Asian” branch are spread in many peoples – Shors, Tubalars, Altai-Kizhi people, Telengits, Sagays, Kyzyl Khakas, Koibals, Teleuts (Y-base) (Kharkov et al., 2009). Hg R1a1a comprises perceptible parts of gene pools of Tuvinian clans (19 % in Mongush, and 15 % in Oorzhak), though its occurrence in Mongols is much lower (6 %). Those results also count in favor of the hypothesis of autochtonous component dominance even in the gene pools of clans potentially most influenced by Mongolian ancestry. If we add R1a1a variants to the “North Eurasian” haplogroups, the “not-Central Asian” component will compose average four fifth of the entire gene pools for Tuvinian clans (in Mongush 77 %, and in Oorzhak 81 %), being only 16 % in Mongols. Such data are definitely contrary to the hypothesis of a crucial influence of the Mongol expansion upon the development of Tuvinian gene pool.
I found interesting the high proportion of R1a-Z93 subclades among Sagays in Khakhasia, which stem from a local Samoyed substratum, as described by the paper…
Featured Image: Map of Uralic and Altaic languages, from Wikipedia.
Related
- Eurasian steppe dominated by Iranian peoples, Indo-Iranian expanded from East Yamna
- Consequences of Damgaard et al. 2018 (III): Proto-Finno-Ugric & Proto-Indo-Iranian in the North Caspian region
- No large-scale steppe migration into Anatolia; early Yamna migrations and MLBA brought LPIE dialects in Asia
- Early Indo-Iranian formed mainly by R1b-Z2103 and R1a-Z93, Corded Ware out of Late PIE-speaking migrations
- Y-DNA haplogroup R1b-Z2103 in Proto-Indo-Iranians?
- North Pontic steppe Eneolithic cultures, and an alternative Indo-Slavonic model
- New Ukraine Eneolithic sample from late Sredni Stog, near homeland of the Corded Ware culture
- The renewed ‘Kurgan model’ of Kristian Kristiansen and the Danish school: “The Indo-European Corded Ware Theory”