This is the second of four posts on the Corded Ware—Uralic identification:
- Corded Ware—Uralic (I): Differences and similarities with Yamna
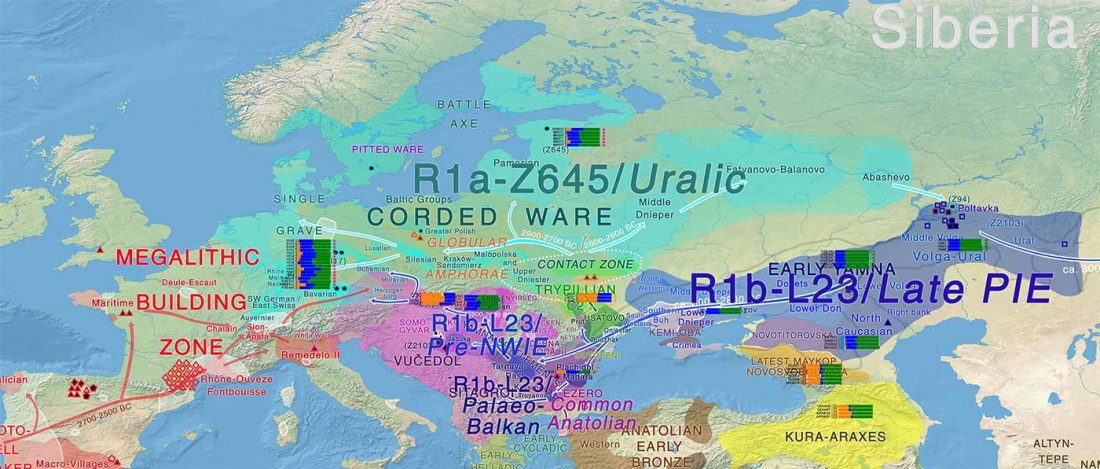
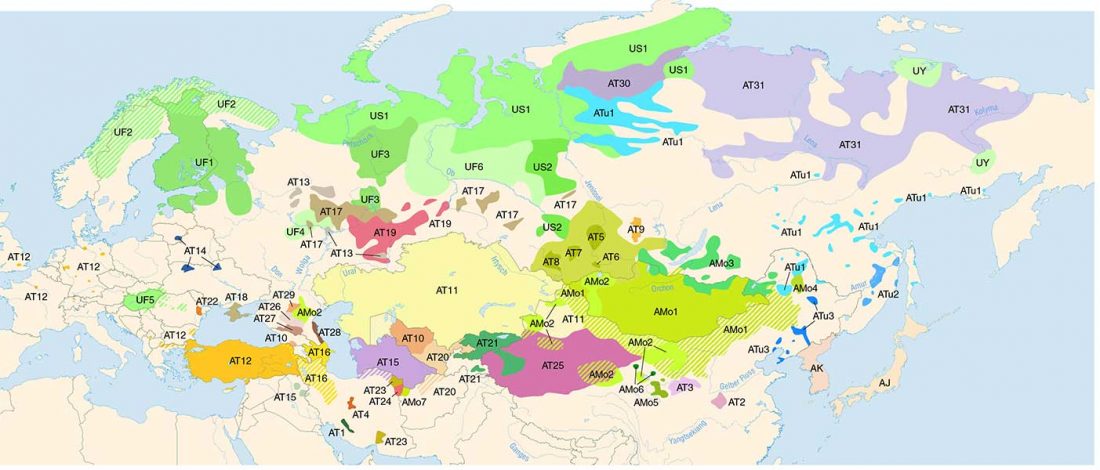
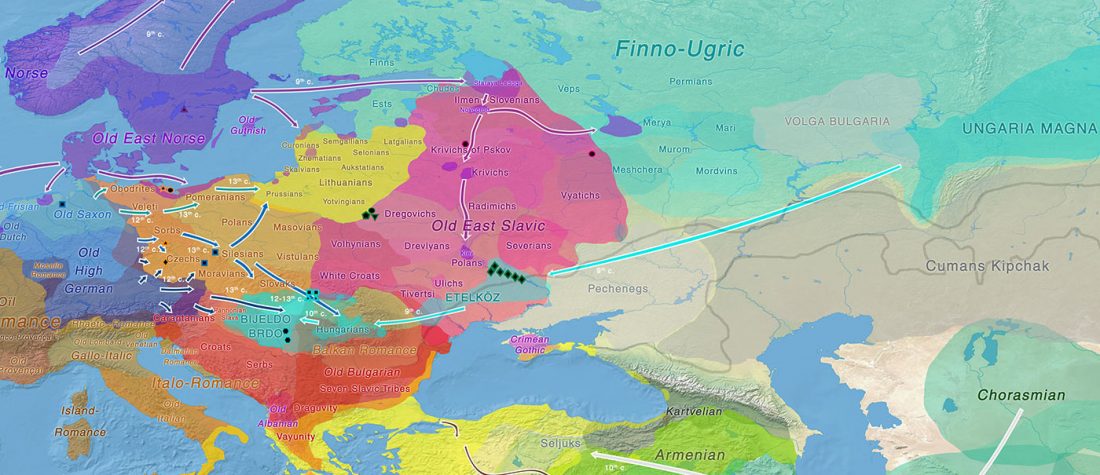
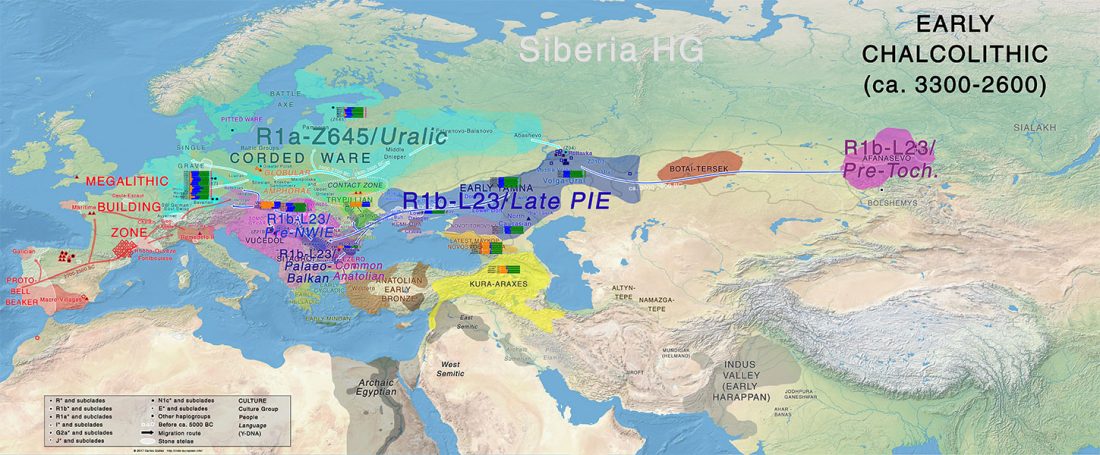
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (IV): Haplogroups R1a and N in Finno-Ugric and Samoyedic
I read from time to time that “we have not sampled Uralic speakers yet”, and “we are waiting to see when Uralic-speaking peoples are sampled”. Are we, though?
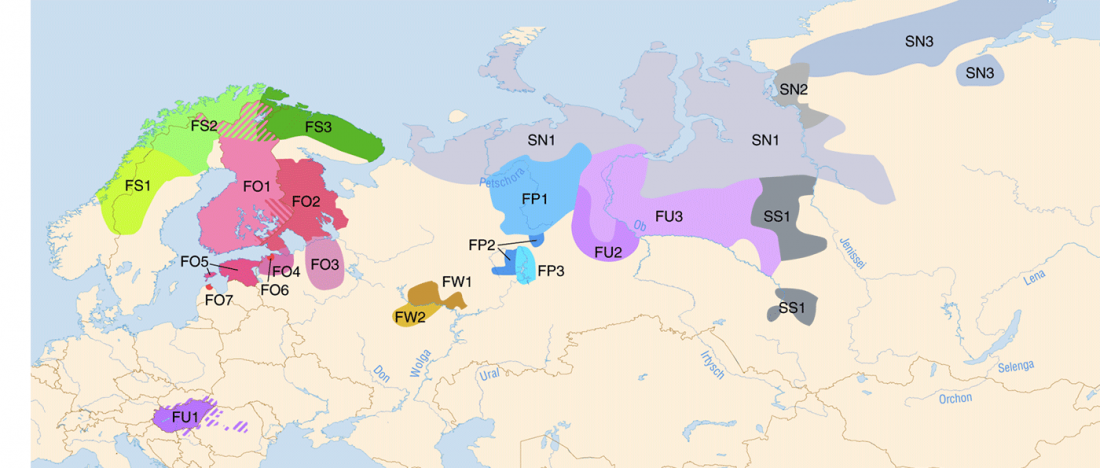
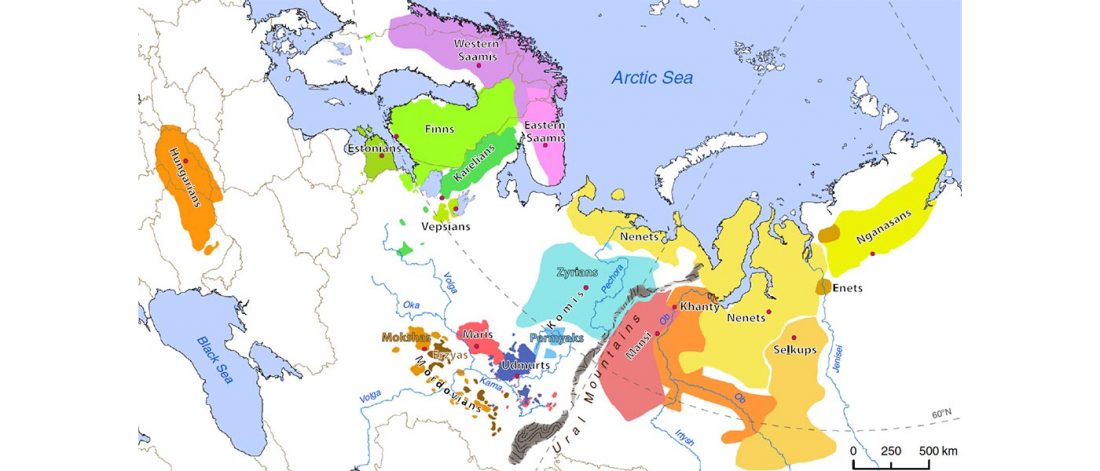
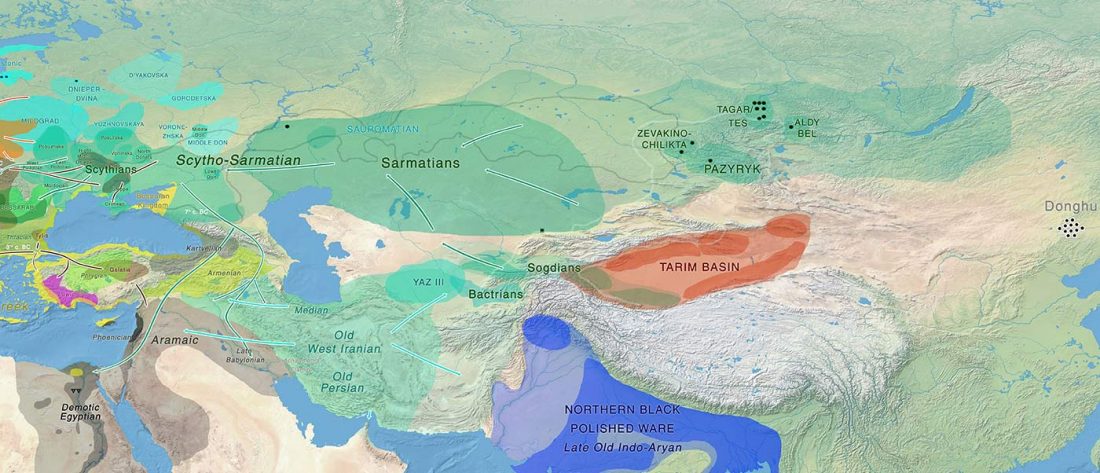
Proto-language homelands are based on linguistic data, such as guesstimates for dialectal evolution, loanwords and phonetic changes for language contacts, toponymy … Read the rest “Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry”