New open access paper from the Max Planck Institute: Analysis of 3800-year-old Yersinia pestis genomes suggests Bronze Age origin for bubonic plague, by Spyrou et al., Nature Communications (2018) 9:2234.
Interesting excerpts from the paper and supplementary materials (emphasis mine):
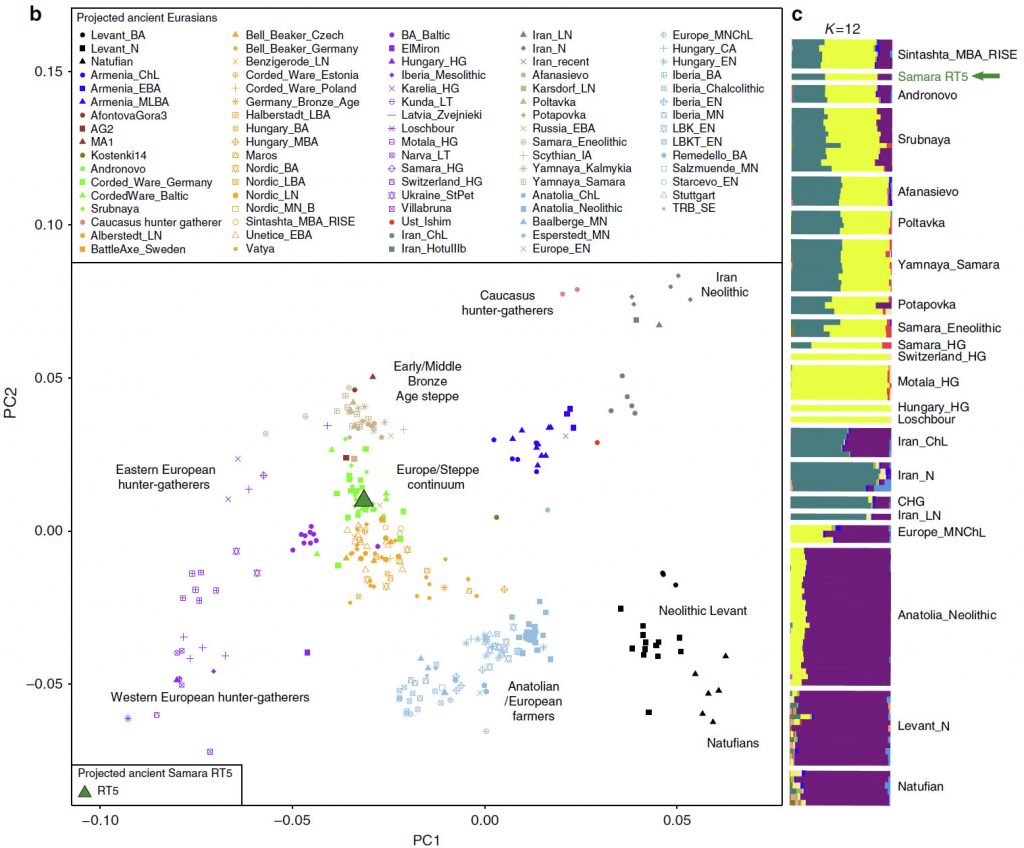
Here, we analyse material from the Mikhailovsky II burial site, which was excavated in 2015 and is one of numerous kurgan cemeteries identified in the Samara Oblast. It consists of seven kurgan burials, and is chronologically associated to the ‘Pokrovka’ phase (3,900-3,750 BP) of the ‘Srubnaya’ culture (3,850-3,150 BP) (radiocarbon dates produced in this study provided in Supplementary Table 6), also referred to as the ‘proto-Srubnaya’ that is considered the earliest phase of the LBA in the Samara Oblast. All sex and age groups were represented in this cemetery. We analysed nine individuals buried in three kurgans and identified two individuals buried in the same kurgan (see Supplementary Figure 1) to be positive for Y. pestis. According to anthropological analysis these were a 30-40 year-old male (RT5) and 35- 45 year-old female (RT6).
After its divergence from Y. pseudotuberculosis, Y. pestis acquired its high pathogenicity and distinct niche mainly by chromosomal gene loss16 as well as the acquisition of two virulence-associated plasmids, pMT1 and pPCP11,17,18. Throughout this process, one of the most crucial evolutionary adaptations related to its pathogenicity was its ability to colonise arthropods, a phenotypic/functional gain mediated by a combination of chromosomal and plasmid loci19,20. These genetic changes are central to the most common “bubonic” form of the disease, where bacteria enter the body via the bite of an infected flea, travel via the lymph to the closest lymph node and replicate while evading host defences. Recent ancient genomic investigations of Y. pestis have identified its earliest known variants in Eurasia during the Late Neolithic/Bronze Age period (LNBA) that show genetic characteristics incompatible with arthropod adaptation. These strains, therefore, have been considered incapable of an efficient flea-based transmission2; however, the alternative early-phase transmission could have provided an independent means of arthropod dissemination2,3,21. To date, the earliest evidence of a Y. pestis strain with signatures associated with flea adaptation has been reported during the Iron Age through shotgun sequencing of an ~2900-year-old genome from Armenia (strain RISE397), though at a coverage too low (0.25-fold) to permit confident phylogenetic positioning2. Although the mechanism by which the LNBA lineage caused human disease is unclear, its frequency in Eurasia during the Bronze Age2,3 and its phylogeographic pattern that mimics contemporaneous human migrations are noteworthy3.
The central steppe region seems to have played a significant role as a migration corridor during the entire Bronze Age, and as such, it likely facilitated the spread of human-associated pathogens, such as Y. pestis, across Eurasia. Here, we explore additional Y. pestis diversity in that region by isolating strains from LBA Samara, in Russia. We identify a Y. pestis lineage contemporaneous to the LNBA strains with genomic variants consistent with flea adaptation. This reveals the co-circulation of two Y. pestis lineages during the Bronze Age with different properties in terms of their transmission and disease potentials.
A recent study has suggested that flea-adapted Y. pestis, along with its potential to cause bubonic plague in humans, likely originated around 3000y BP2. Contrary to such conclusions, the lineage giving rise to our Y. pestis isolates (RT5 and RT6) likely arose ~4000 years ago (Supplementary Tables 6 and 9), and possessed all vital genetic characteristics required for flea-borne transmission of plague in rodents, humans and other mammals. (…)
Moreover, our analysis of the previously published Iron Age RISE397 strain from modern-day Armenia2 revealed its close relationship to RT5 and RT6 (Supplementary Fig. 4). Note that the modern 0.PE2 and 0.PE7 lineages, which are known to possess all genomic characteristics that confer adaptation to fleas19, fall ancestral to RT5 (Fig. 2b) and RISE397 (Supplementary Fig. 4), but are more derived than the LNBA lineage. Our phylogenetic and dating results thus suggest that 0.PE2 and 0.PE7 also originated during the Bronze Age, with their mean divergence here estimated to 4474 (HPD 95%: 3936–5158) and 5237 (HPD 95%: 4248–6346) years BP, respectively, based on the Bayesian skyline model (Supplementary Table 9). While these lineages may have been confined to sylvatic rodent reservoirs during the EBA, the possibility that they co circulated among human populations contemporaneously with the LNBA lineage should be considered. Although the places of origin of 0.PE2 and 0.PE7 are not known, today, their strains are isolated from modern-day China and the Caucasus region. In terms of their disease potential, both 0.PE2 and 0.PE7 possess pMT1 plasmids with fully functional ymt genes, but 0.PE2 strains lack pPCP144, and though frequently recovered from sylvatic rodent reservoirs, their virulence in humans is not known. On the other hand, the more basal 0.PE7 contains pPCP12 and has previously been associated with human bubonic plague12. It is, therefore, tempting to hypothesise that efficient flea adaptation in Y. pestis, as well as the potential for bubonic disease, might have evolved earlier than 5000 years ago.
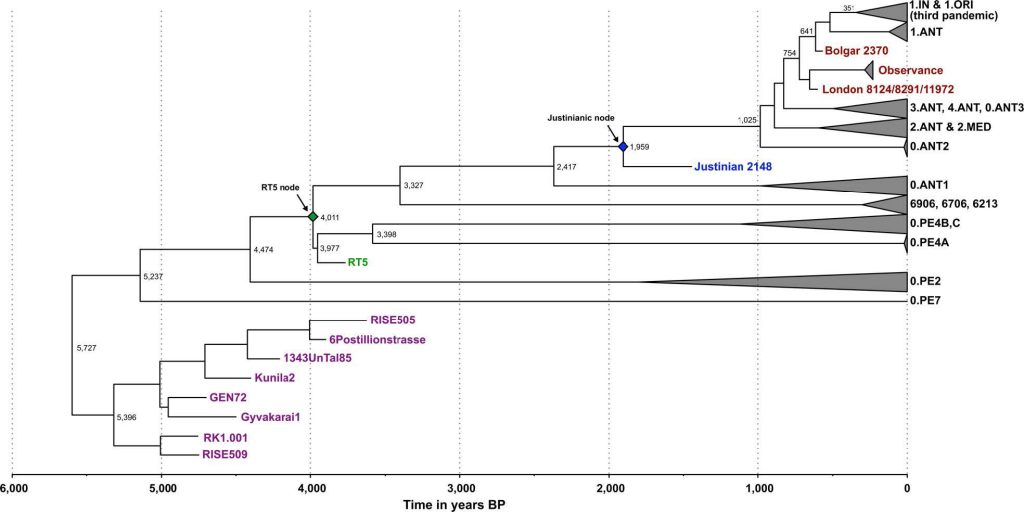
It seems possible that already in the Bronze Age, with the establishment of transport and trade networks, the interconnectivity between Europe and Asia that is also reflected in the ancient human genomes, likely contributed to the spread of infectious disease. Similarly, the abundant trade routes of the medieval period are considered the main conduit for plague’s movement between Asia and Europe8,12. Our current data suggest a more complex model, where at least two human-associated lineages (LNBA and RT5) with different transmission potentials were established in Eurasia during the Bronze Age (Fig. 2b, c).
The haplogroup of RT5 is R1a1a1b-Z645 (most likely Z93, only with coverage of 1-fold), mtDNA U2e2a.
See also materials from the Max Plank Institute.
Related:
- Phylogeny of leprosy, relevant for prehistoric Eurasian contacts
- Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans
- Stone Age plague accompanying migrants from the steppe, probably Yamna, Balkan EBA, and Bell Beaker, not Corded Ware
- The Caucasus a genetic and cultural barrier; Yamna dominated by R1b-M269; Yamna settlers in Hungary cluster with Yamna
- Steppe and Caucasus Eneolithic: the new keystones of the EHG-CHG-ANE ancestry in steppe groups
- East Bell Beakers, an in situ admixture of Yamna settlers and GAC-like groups in Hungary