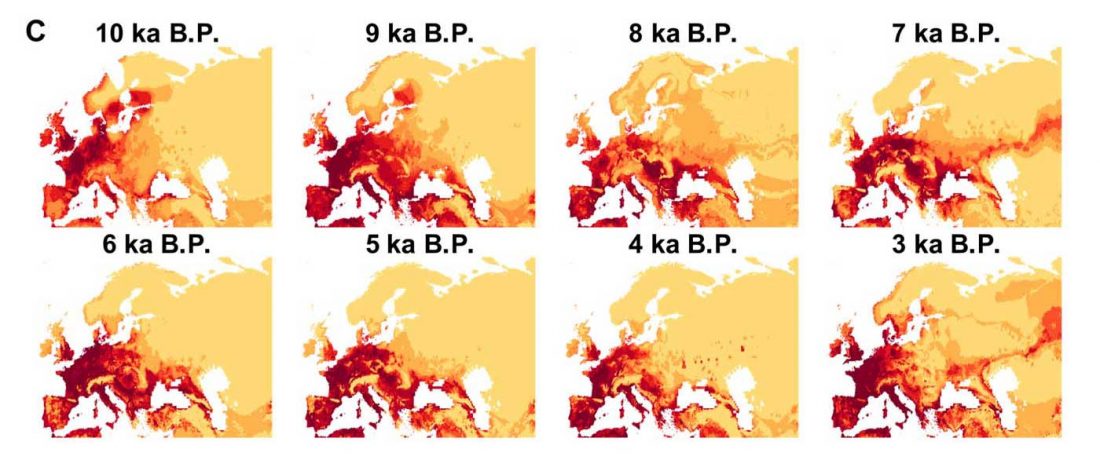
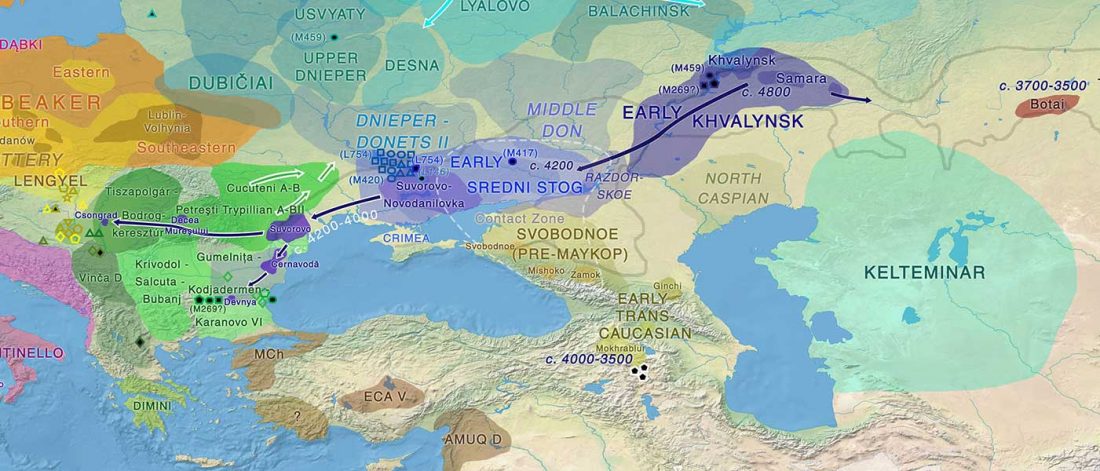
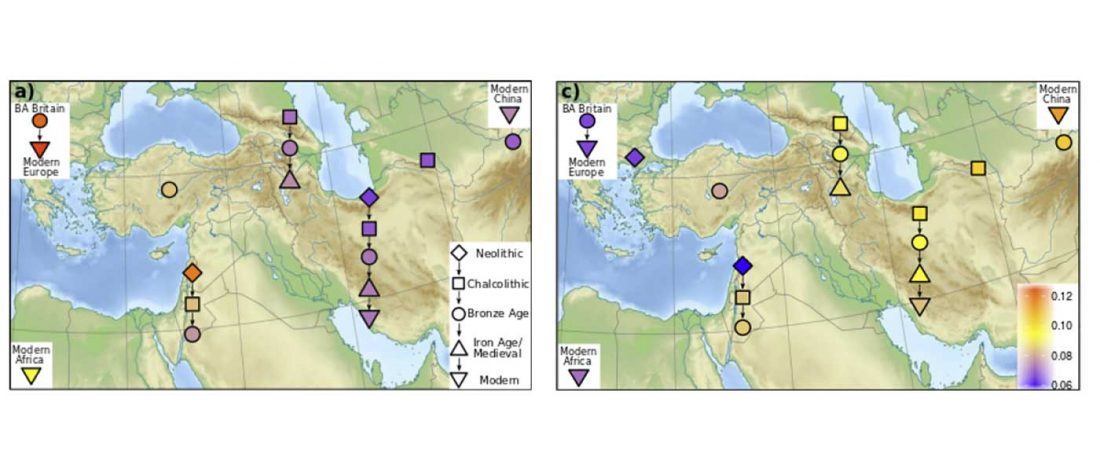
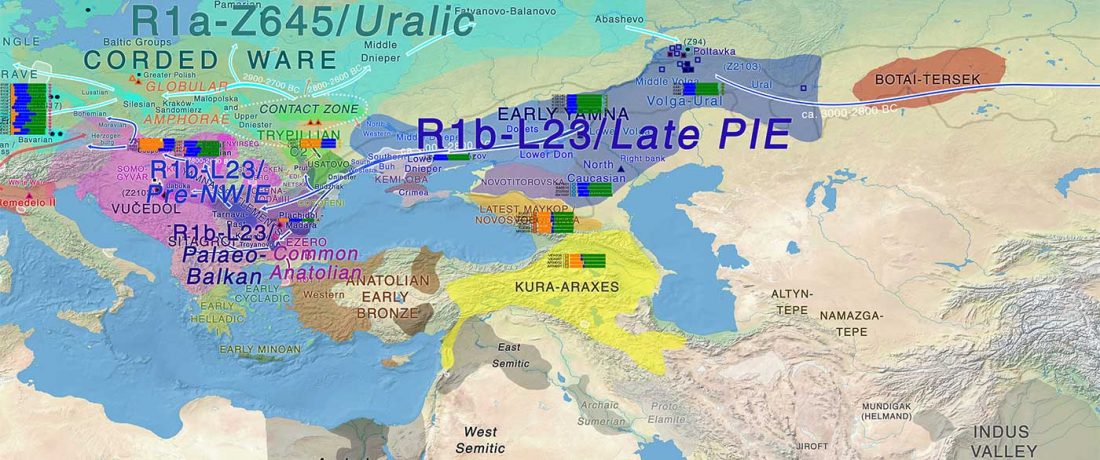
New preprint Population history from the Neolithic to present on the Mediterranean island of Sardinia: An ancient DNA perspective, by Marcus et al. bioRxiv (2019)
Interesting excerpts (emphasis mine, edited for clarity):
On the high frequency of R1b-V88
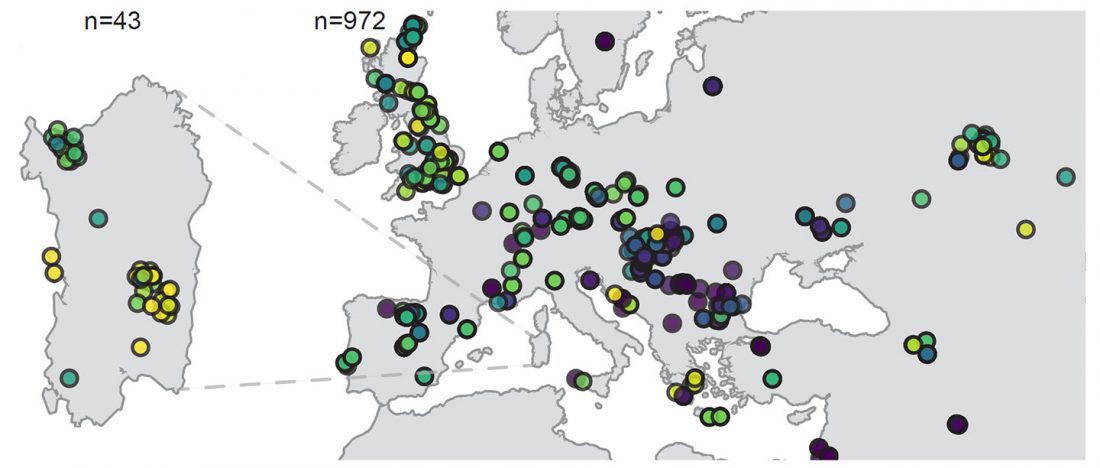
… Read the rest “Ancient Sardinia hints at Mesolithic spread of R1b-V88, and Western EEF-related expansion of Vasconic”Our genome-wide data allowed us to assign Y haplogroups for 25 ancient Sardinian individuals. More than half of them consist of R1b-V88 (n=10) or I2-M223 (n=7).
Francalacci et al. (2013) identified three major Sardinia-specific founder clades based on present-day variation within the haplogroups I2-M26, G2-L91 and R1b-V88, and here we found each of those broader haplogroups in at least one