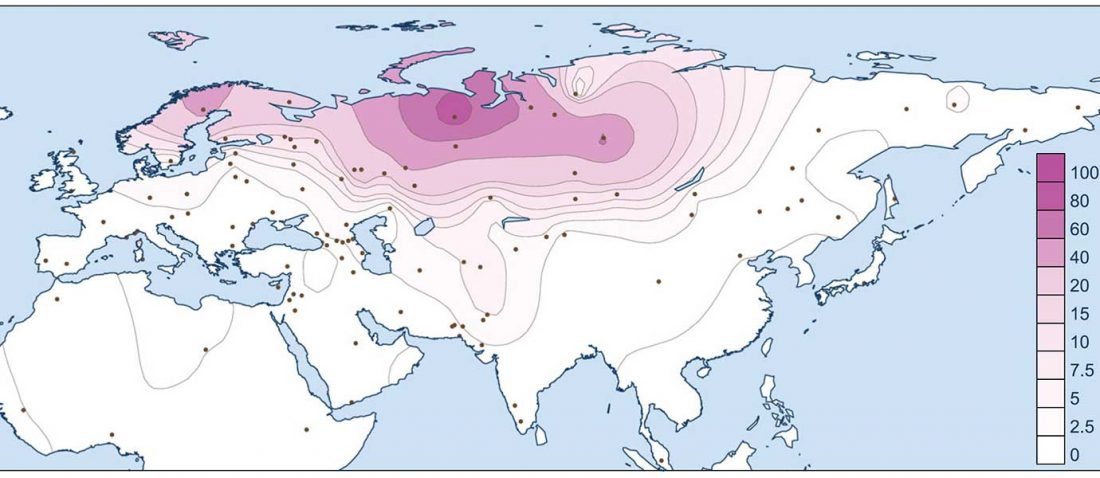
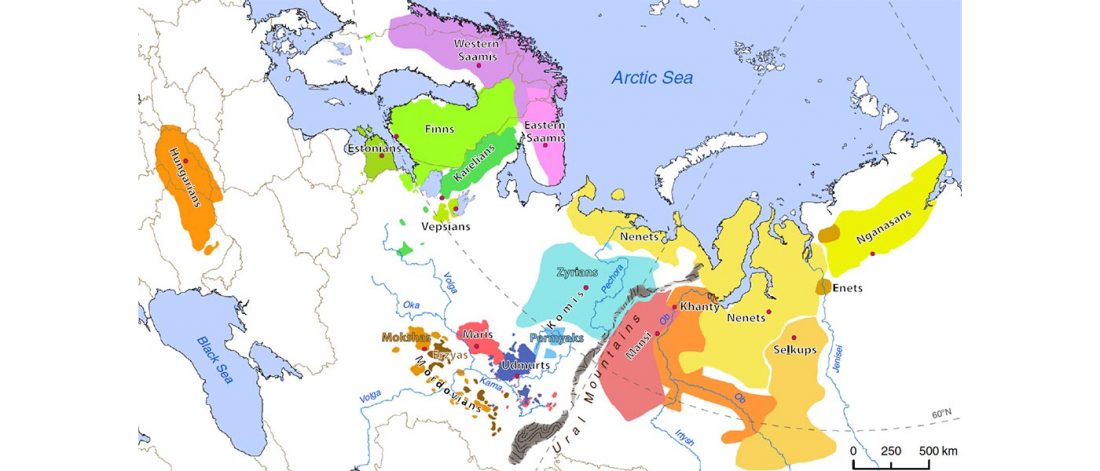
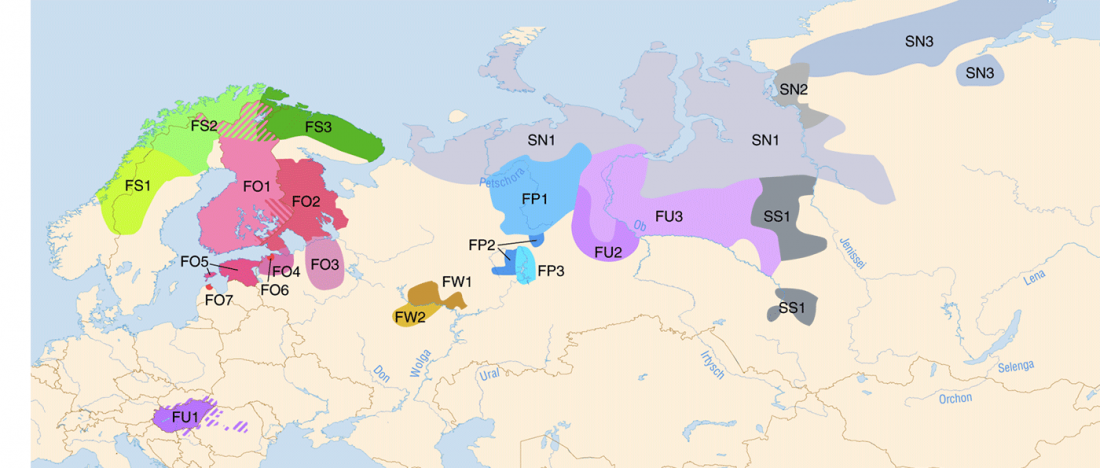
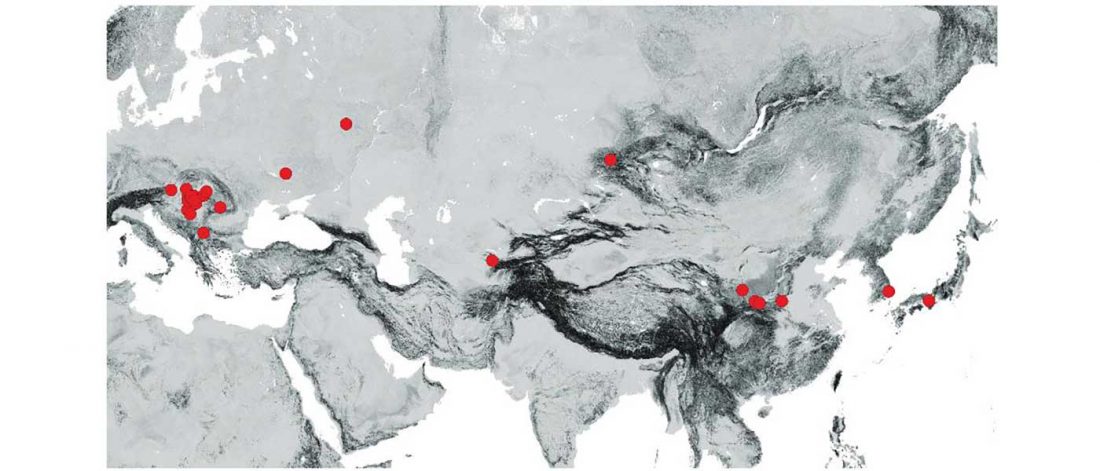
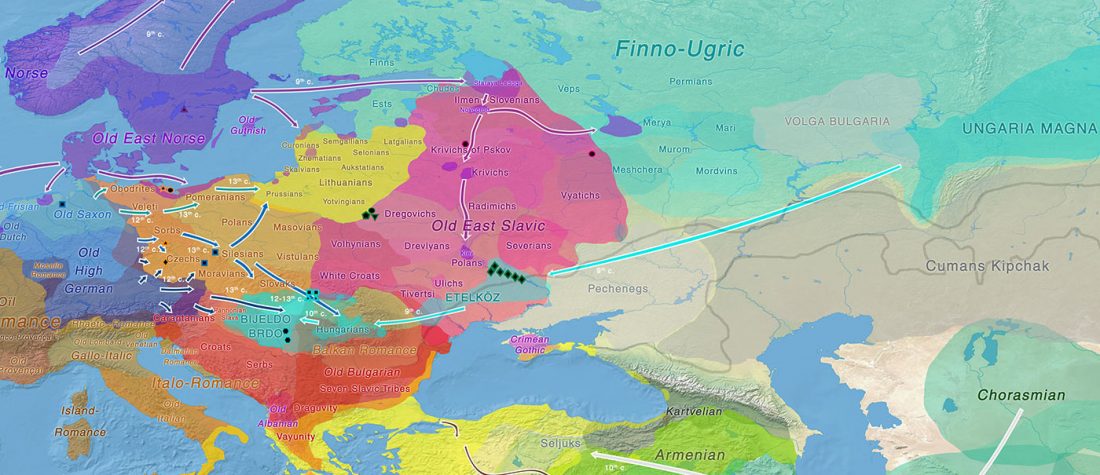
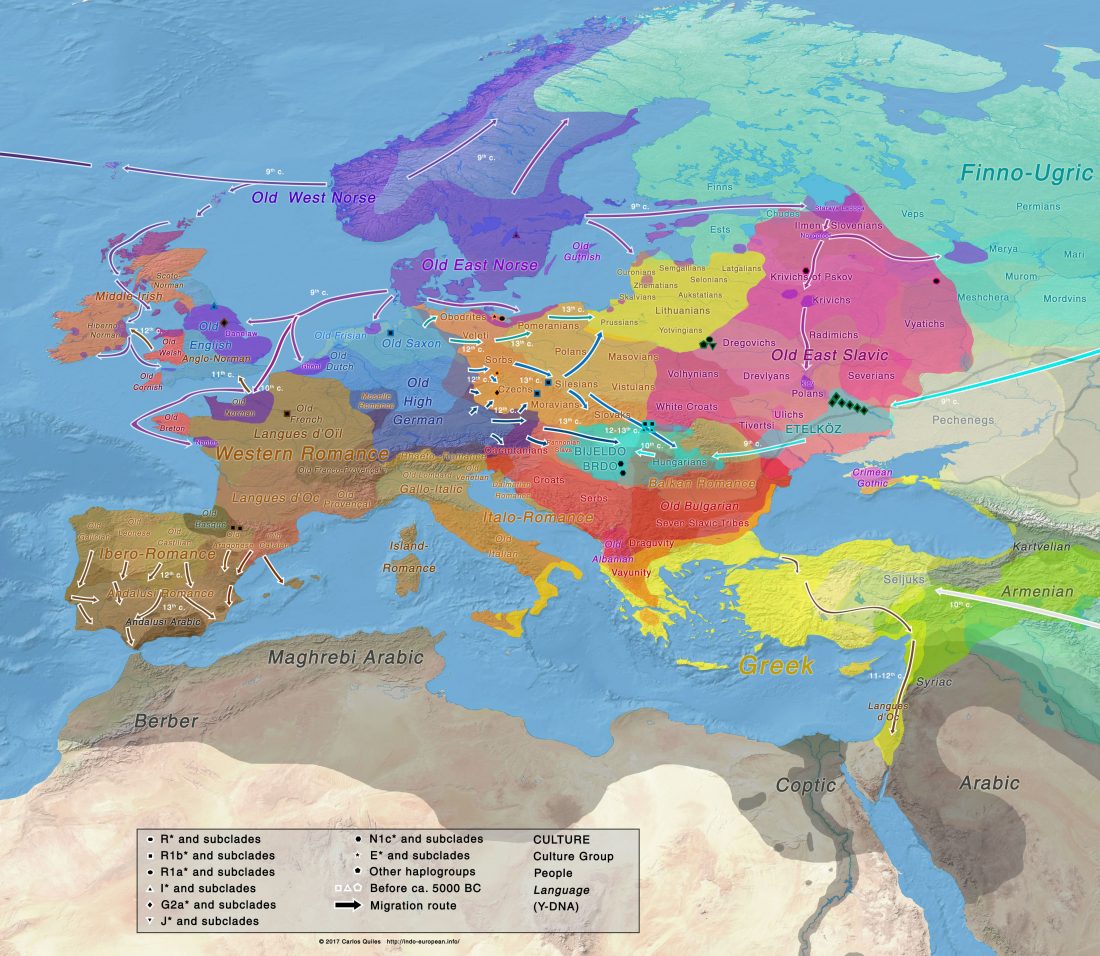
Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
This is the third of four posts on the Corded Ware—Uralic identification. See
- Corded Ware—Uralic (I): Differences and similarities with Yamna
- Corded Ware—Uralic (II): Finno-Permic and the expansion of N-L392/Siberian ancestry
- Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions
- Corded Ware—Uralic (IV): Haplogroups R1a and N in Finno-Ugric and Samoyedic
An Eastern Uralic group?
Even though proposals of an Eastern Uralic (or Ugro-Samoyedic) group are in the minority – and those who support it tend to search for an origin of Uralic in Central Asia – , there is nothing wrong in supporting this from the point of view … Read the rest “Corded Ware—Uralic (III): “Siberian ancestry” and Ugric-Samoyedic expansions”