Inner Asian maternal genetic origin of the Avar period nomadic elite in the 7th century AD Carpathian Basin, by Csáky et al. bioRxiv (2018).
Abstract (emphasis mine):
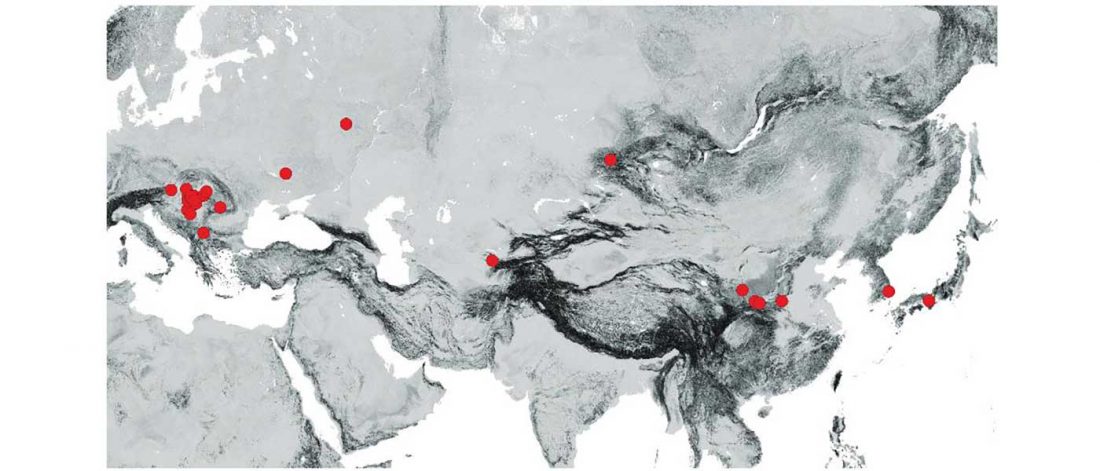
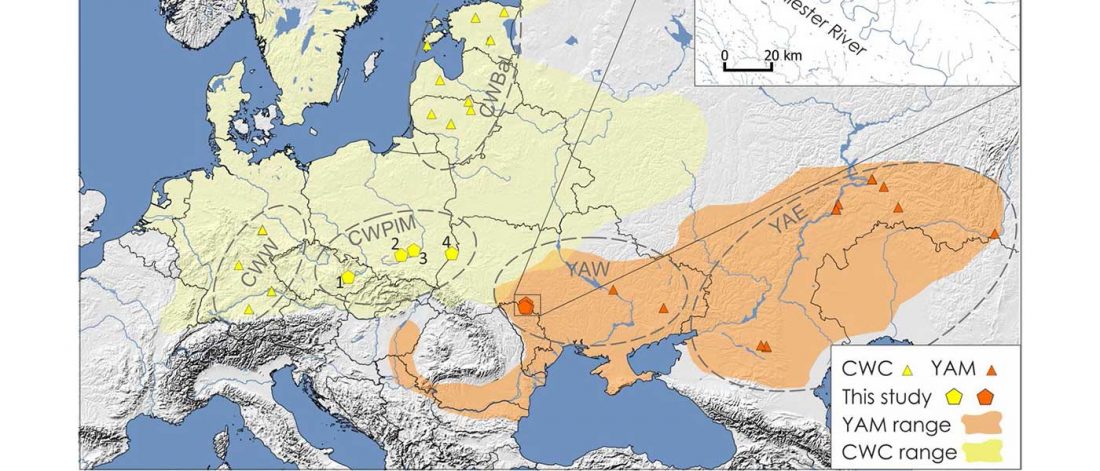
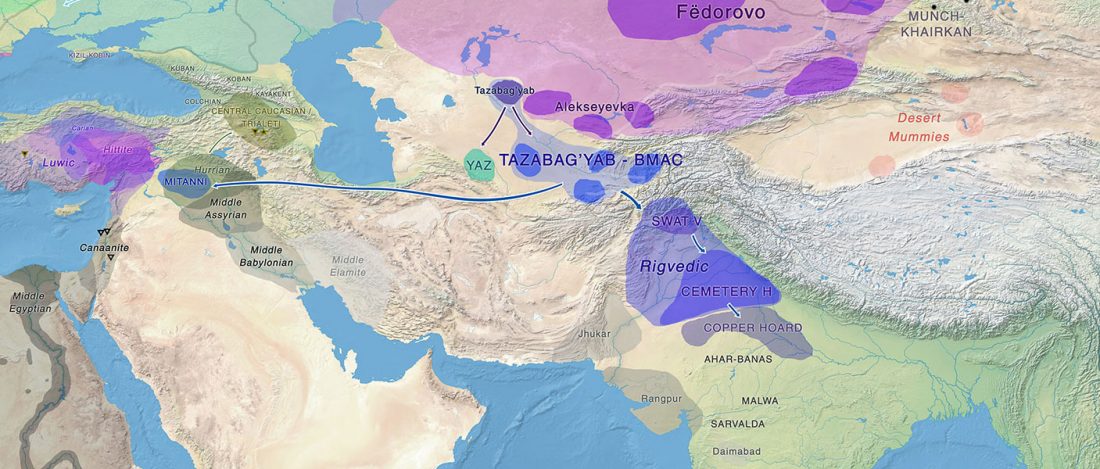
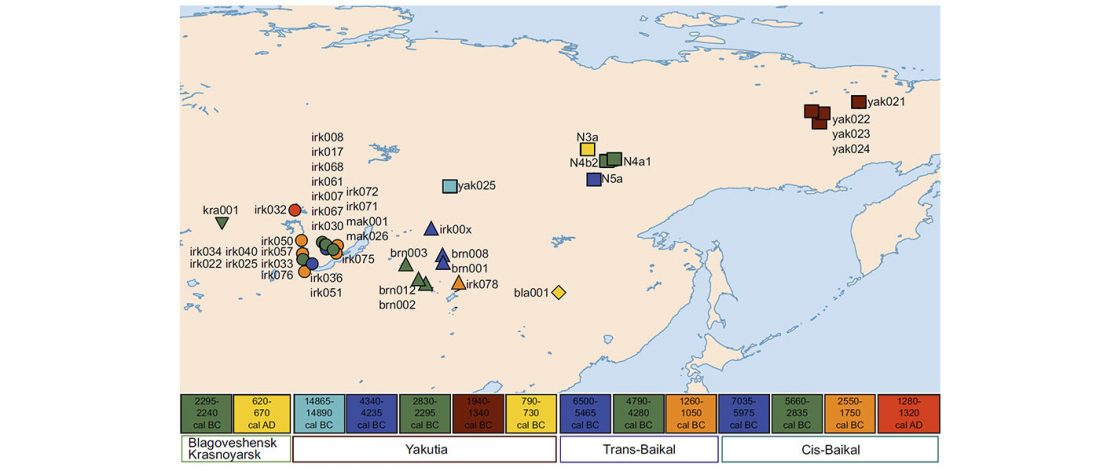
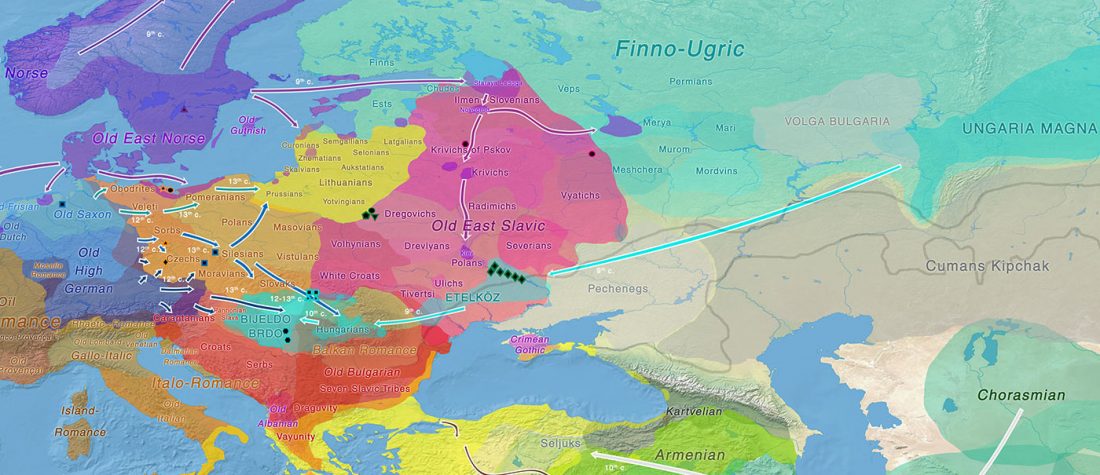
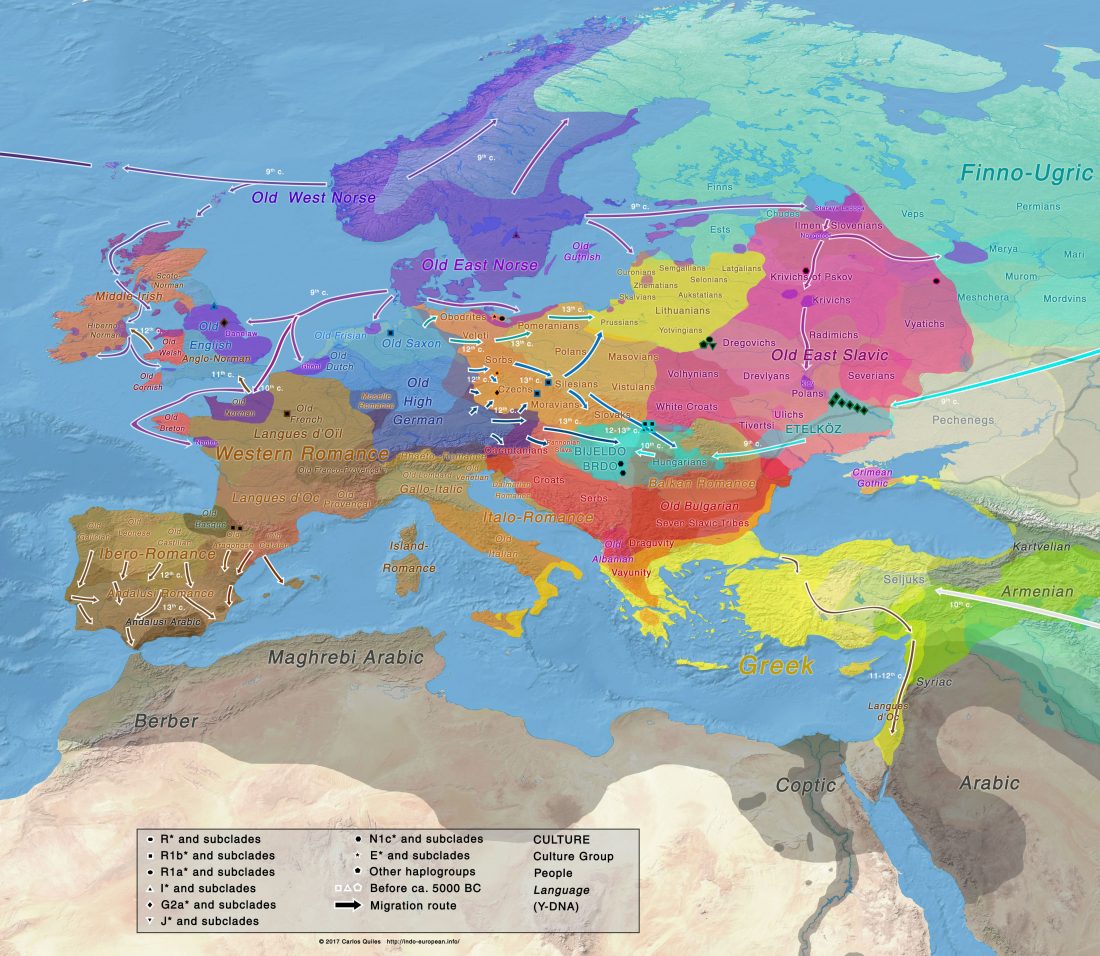
… Read the rest “Mitogenomes from Avar nomadic elite show Inner Asian origin”After 568 AD the nomadic Avars settled in the Carpathian Basin and founded their empire, which was an important force in Central Europe until the beginning of the 9th century AD. The Avar elite was probably of Inner Asian origin; its identification with the Rourans (who ruled the region of today’s Mongolia and North China in the 4th-6th centuries AD) is widely accepted in the historical research.
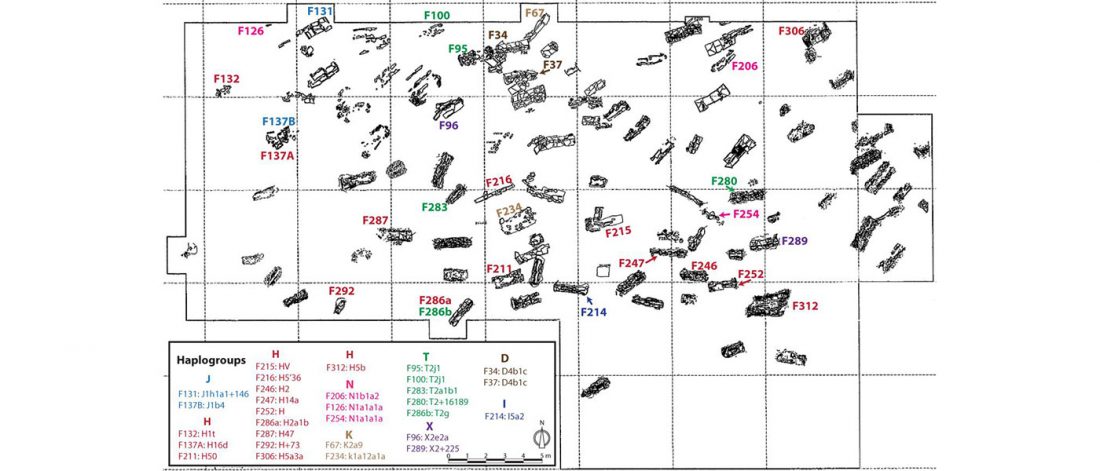
Here, we study the whole