Another interesting finding from Human auditory ossicles as an alternative optimal source of ancient DNA, by Sirak et al. (2020):
A sample classified as Italy Middle Bronze Age from Olmo di Nogara (ca. 1400-1200 BC), who is R1b-L51 (xP311, xL52, xL151), CTS6889+ (T->C, 1 read). See YFull’s corresponding R-S1161.
This sample probably belongs to individual 309 (35-45 yo), and the female sampled to 323 (30-40 yo), both referenced as from the following study:
Canci A, Contursi D, Fornaciari G. 2005. La necropoli dell’età del bronzo di Olmo di Nogara (Verona): primi risultati
…
Read the rest “Italo-Venetic peoples related patrilineally to Terramare elites”
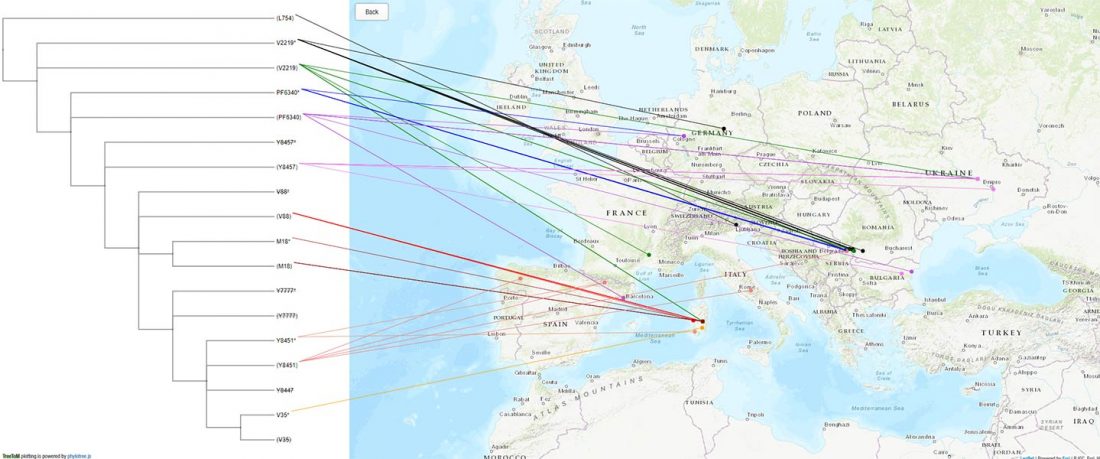
Yesterday the Eaton Lab at Columbia University announced on Twitter a nifty little tool by Carlos Alonso Maya-Lastra called TreeToM, which accepts Newick trees and CSV latitude/longitude data to explore phylogeny and geography interactively, with no coding required.
I thought it could complement nicely my All Ancient DNA Dataset, particularly for those newly described SNPs (FTDNA private variants, etc.) that have not been incorporated yet into SNP Tracker.
Here are two examples with snippets to copy&paste to the appropriate boxes in TreeToM. Feel free to add others in the comments:… Read the rest “Visualizing phylogenetic trees of ancient DNA in a map”
New paper (behind paywall) Ancient Rome: A genetic crossroads of Europe and the Mediterranean, by Antonio et al. Science (2019).
The paper offers a lot of interesting data concerning the Roman Empire and more recent periods, but I will focus on Italic and Etruscan origins.
NOTE. I have updated prehistoric maps with Y-DNA and mtDNA data, and also the PCA of ancient Eurasian samples by period including the recently published samples, now with added sample names to find them easily by searching the PDFs.
Apennine homeland problem
The traditional question of Italic vs. Etruscan origins from a cultural-historical … Read the rest “R1b-L23-rich Bell Beaker-derived Italic peoples from the West vs. Etruscans from the East”
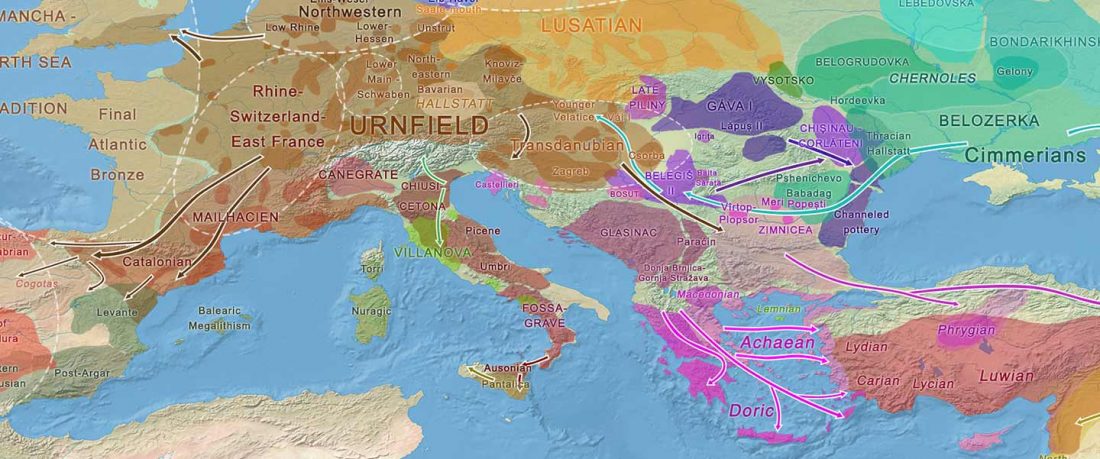
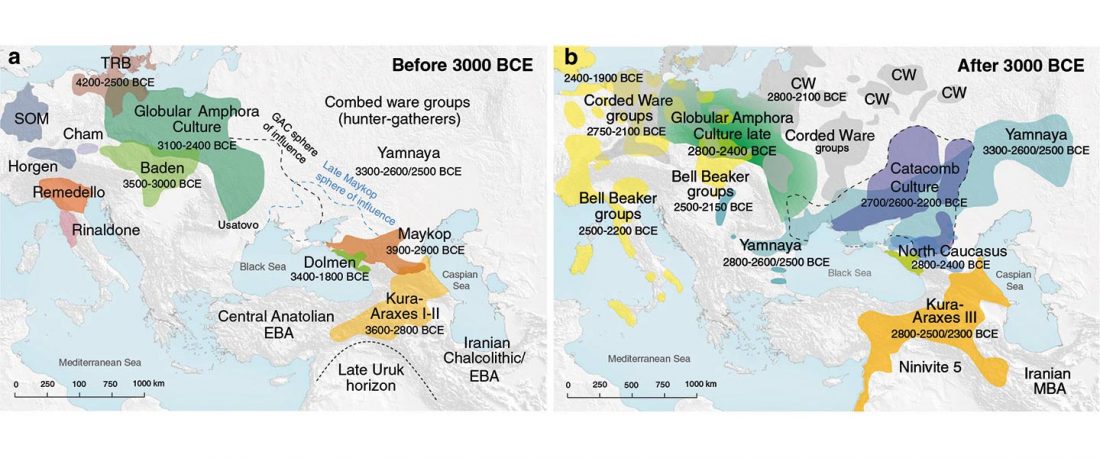
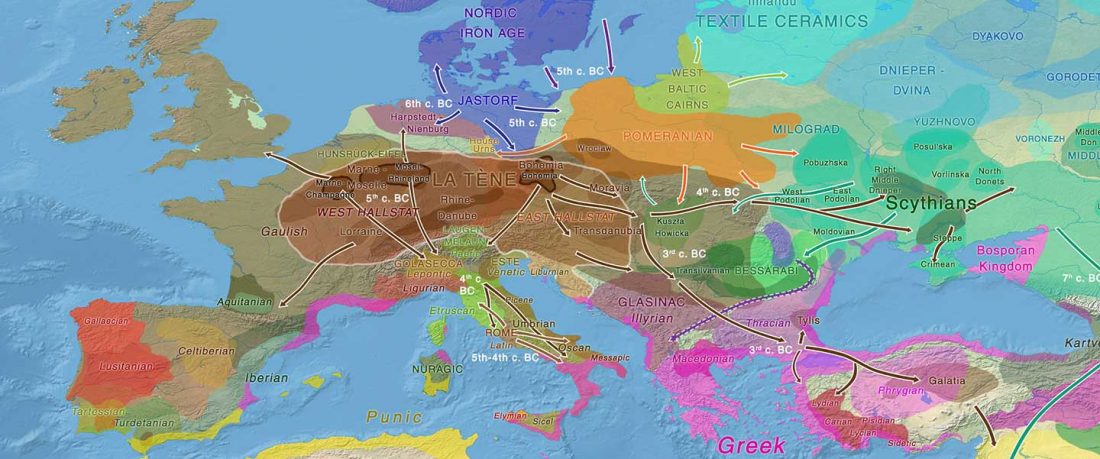
The recent update on the Indo-Anatolian homeland in the Middle Volga region and its evolution as the Indo-Tocharian homeland in the Don–Volga area as described in Anthony (2019) has, at last, a strong scientific foundation, as it relies on previous linguistic and archaeological theories, now coupled with ancient phylogeography and genomic ancestry.
There are still some inconsistencies in the interpretation of the so-called “Steppe ancestry”, though, despite the one and a half years that have passed since we first had access to the closest Pontic–Caspian steppe source populations. Even my post “Steppe ancestry” step by step from a year ago … Read the rest ““Steppe ancestry” step by step (2019): Mesolithic to Early Bronze Age Eurasia”
The recent data on ancient DNA from Iberia published by Olalde et al. (2019) was interesting for many different reasons, but I still have the impression that the authors – and consequently many readers – focused on not-so-relevant information about more recent population movements, or even highlighted the least interesting details related to historical events.
I have already written about the relevance of its findings for the Indo-European question in an initial assessment, then in a more detailed post about its consequences, then about the arrival of Celtic languages with hg. R1b-M167, and later in combination with … Read the rest “North-West Indo-Europeans of Iberian Beaker descent and haplogroup R1b-P312”
Interesting recent developments:
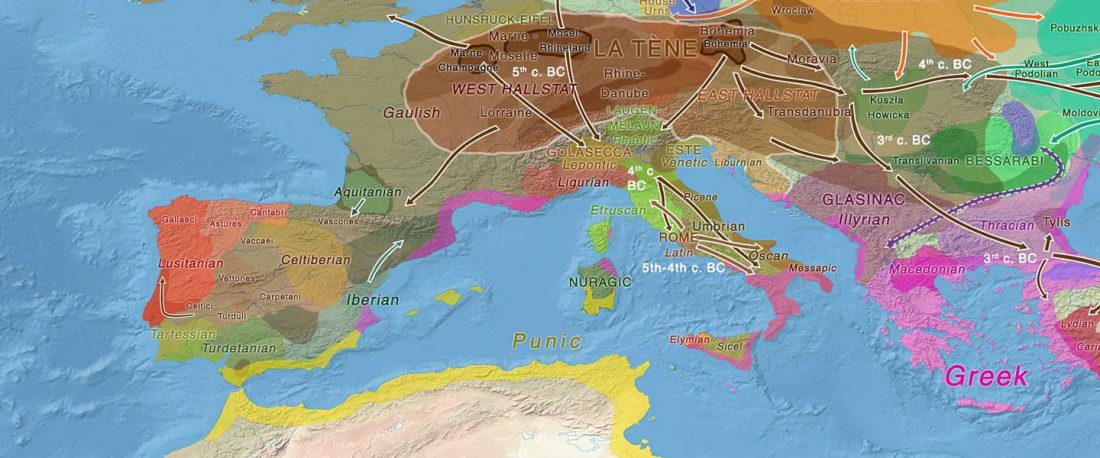
Celts and hg. R1b
Gauls
Recent paper (behind paywall) Multi-scale archaeogenetic study of two French Iron Age communities: From internal social- to broad-scale population dynamics, by Fischer et al. J Archaeol Sci (2019).
In it, Fischer and colleagues update their previous data for the Y-DNA of Gauls from the Urville-Nacqueville necropolis, Normandy (ca. 300-100 BC), with 8 samples of hg. R, at least 5 of them R1b. They also report new data from the Gallic cemetery at Gurgy ‘Les Noisats’, Southern Paris Basin (ca. 120-80 BC), with 19 samples of hg. R, at least 13 of … Read the rest “More Celts of hg. R1b, more Afanasievo ancestry, more maps”
These are maps of modern distribution of haplogroup E-M96 and its main subclades, using natural neighbour interpolation.
Data mainly from D’Atanasio et al. (2018).
E1b-V13
Haplogroup E1b-V13 was probably associated with the expansion of European groups since the Bronze Age, and has been found in ancient DNA among expanding East Germanic and early Slavic peoples.
Pending: other E subclades.… Read the rest “Haplogroup E-M96”
These are maps of modern distribution of ‘archaic’ subclades of haplogroup R-M207, not belonging to R1b-M269 or R1a-M417.
R1-M173
R1a-M420
R1b-M343
Data mainly from Myres et al. (2011).
R1b-L388
R1b-M73
Data mainly from Myres et al. (2011) and various recent papers on R1b-M73 in Asian populations.
R1b-V88
Data mainly from D’Atanasio et al. (2018).
R2-M479
Pending: compile data for mapping of modern distribution of hg. R2.… Read the rest “Haplogroup R1-M173, R2-M479”
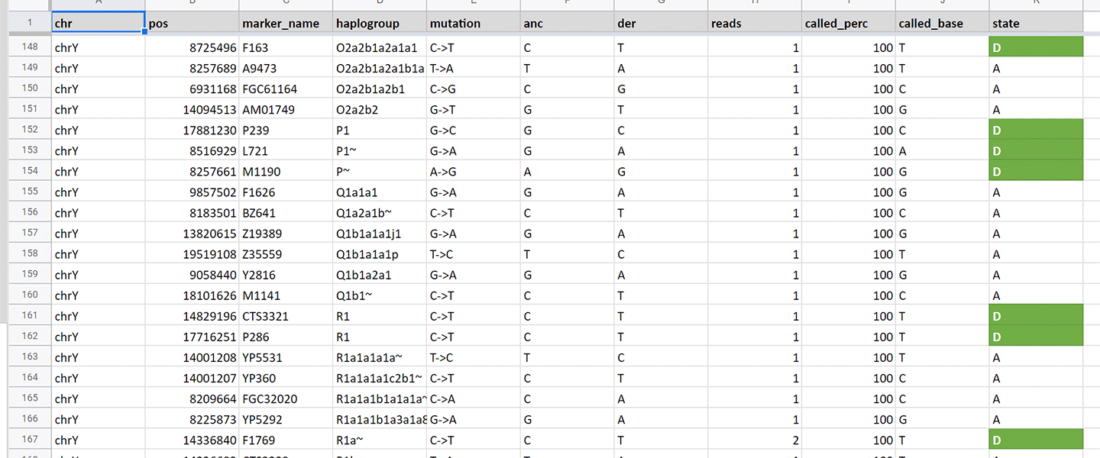
This page hosts analyses of ancient human Y-chromosomal haplogroup inference from next generation sequencing data, performed with software YLeaf v.2. For information on data and interpretation, refer to the manual at Erasmus MC Resources.
As a quick reference on data interpretation, please read papers on ancient DNA damage, such as Dabney, Meyer, and Pääbo (2013), for information of which derivative (and ancestral) SNP calls may be wrong.
For the most recent automated Y-SNP calls obtained with Yleaf v.2.3 or later over FASTQ files, including discussion of individual samples, see:
These are recent … Read the rest “Ancient Y-DNA haplogroups”