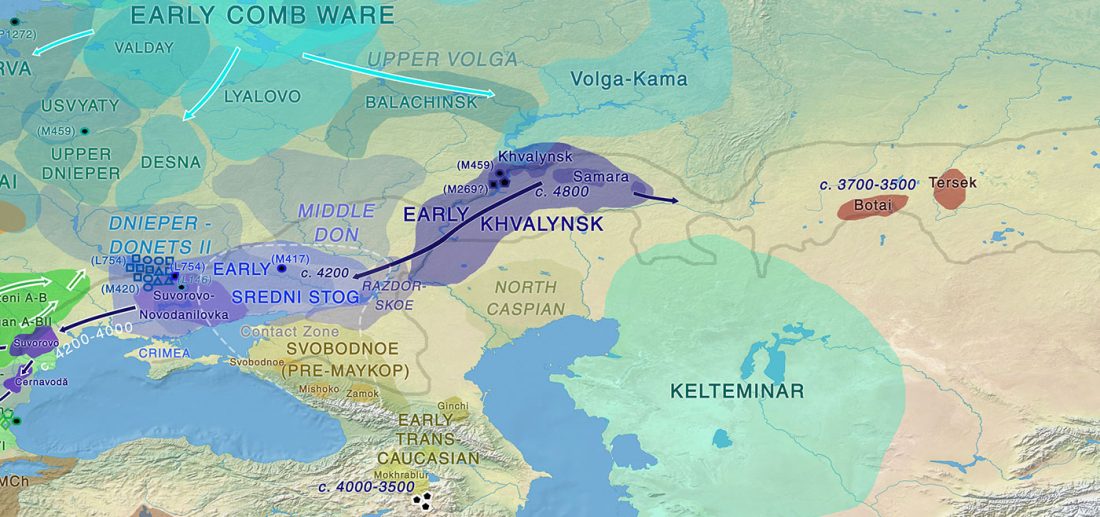
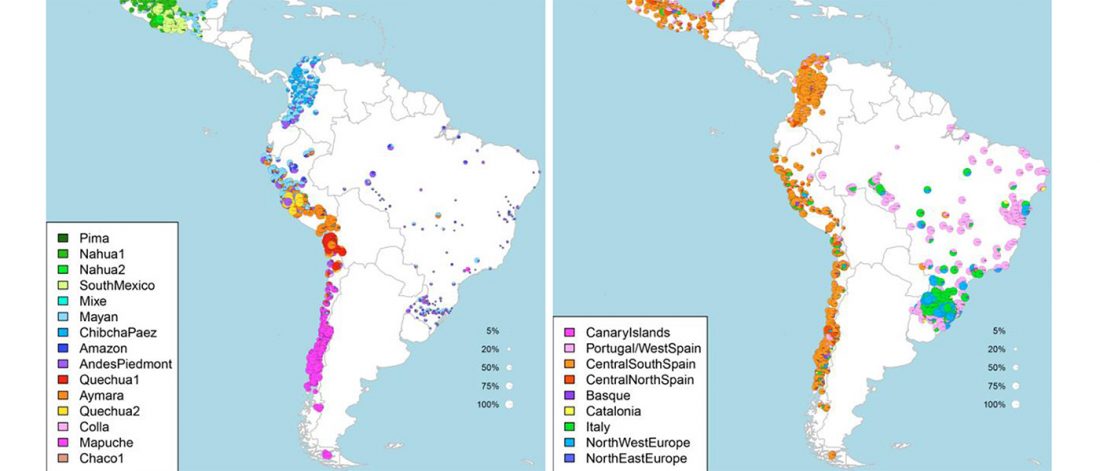
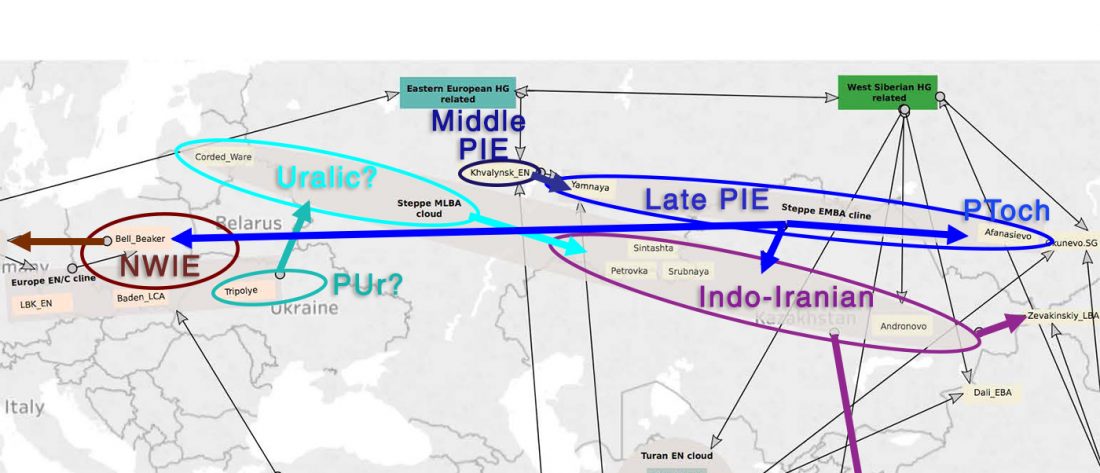
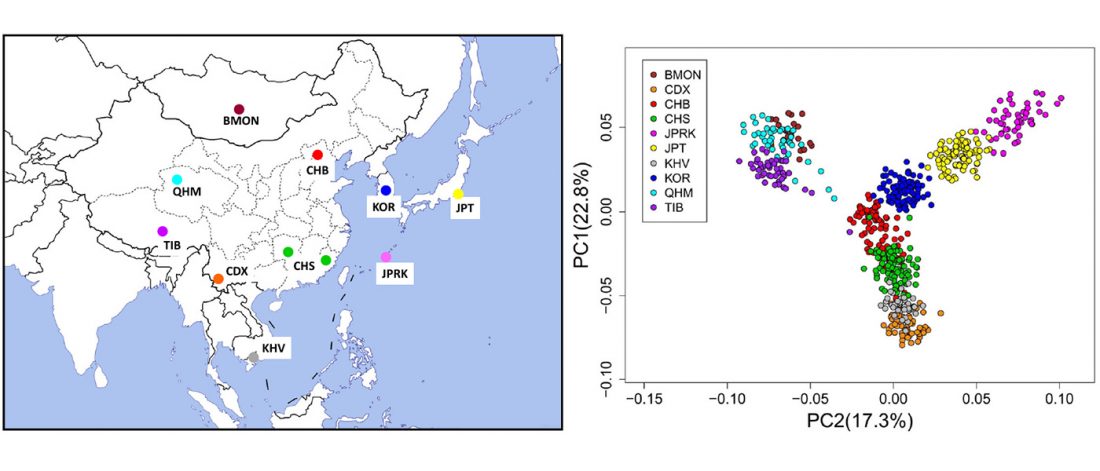
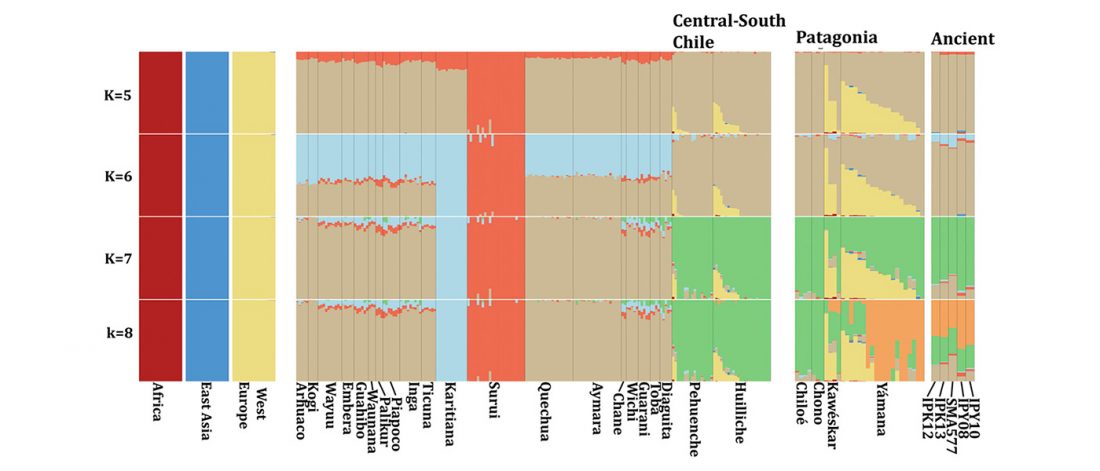
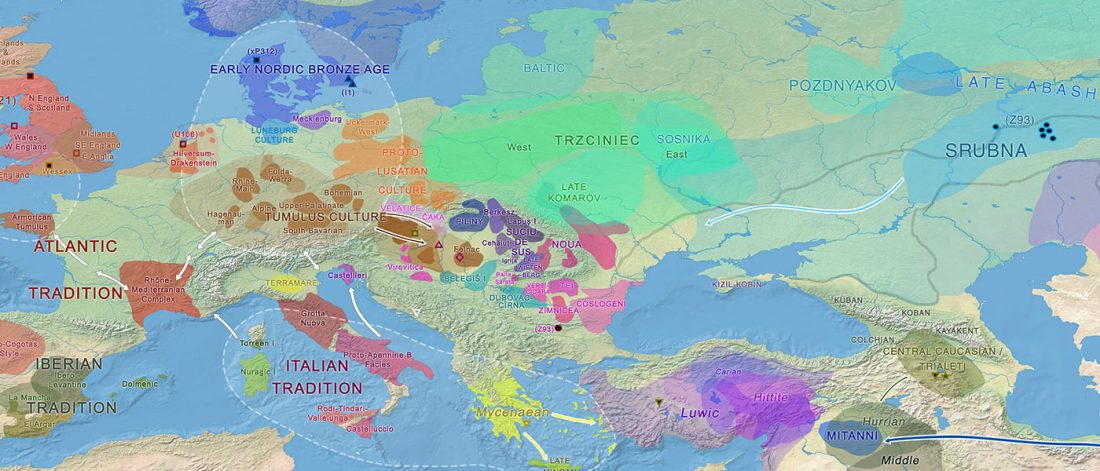
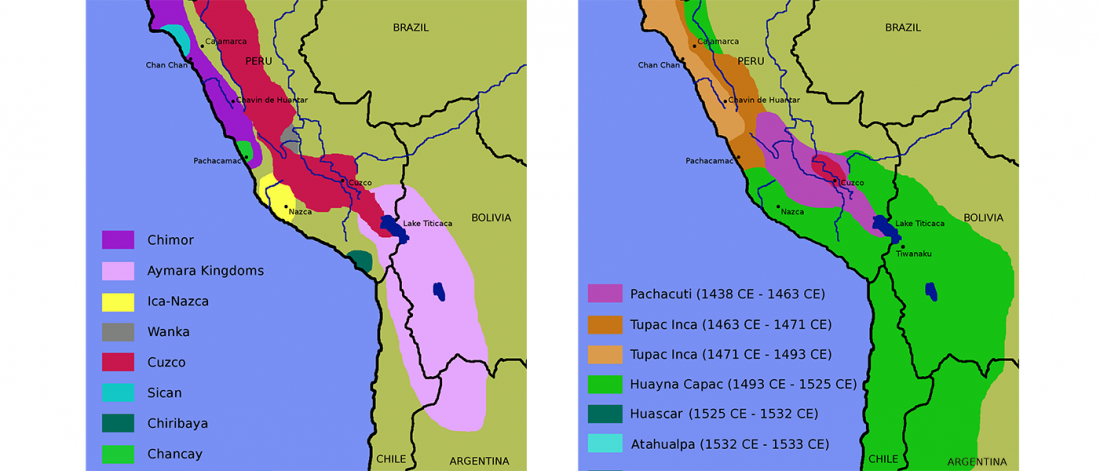
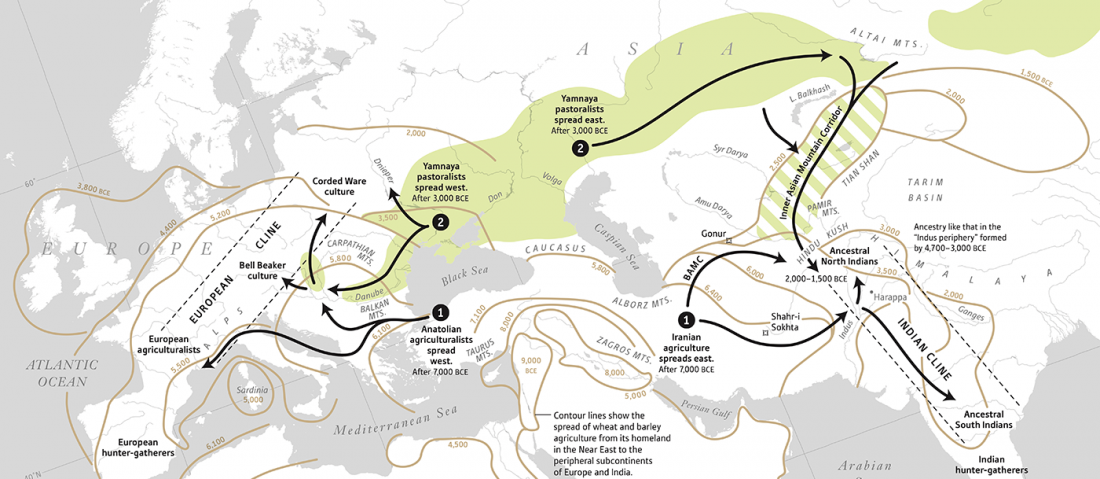
Decline of genetic diversity in ancient domestic stallions in Europe
Open access research article Decline of genetic diversity in ancient domestic stallions in Europe, by Wutke et al., Science (2018), 4(4):eaap9691.
Abstract (emphasis mine):
… Read the rest “Decline of genetic diversity in ancient domestic stallions in Europe”Present-day domestic horses are immensely diverse in their maternally inherited mitochondrial DNA, yet they show very little variation on their paternally inherited Y chromosome. Although it has recently been shown that Y chromosomal diversity in domestic horses was higher at least until the Iron Age, when and why this diversity disappeared remain controversial questions. We genotyped 16 recently discovered Y chromosomal single-nucleotide polymorphisms in 96 ancient Eurasian stallions spanning the early domestication stages (Copper and