New paper (behind paywall) The prehistoric peopling of Southeast Asia, by McColl et al. (Science 2018) 361(6397):88-92 from a recent bioRxiv preprint.
Interesting is this apparently newly reported information including a female sample from the Ikawazu Jōmon of Japan ca. 570 BC (emphasis mine):
The two oldest samples — Hòabìnhians from Pha Faen, Laos [La368; 7950 with 7795 calendar years before the present (cal B.P.)] and Gua Cha, Malaysia (Ma911; 4415 to 4160 cal B.P.)—henceforth labeled “group 1,” cluster most closely with present-day Önge from the Andaman Islands and away from other East Asian and Southeast-Asian populations (Fig. 2), a pattern that differentiates them from all other ancient samples. We used ADMIXTURE (14) and fastNGSadmix (15) to model ancient genomes as mixtures of latent ancestry components (11). Group 1 individuals differ from the other Southeast Asian ancient samples in containing components shared with the supposed descendants of the Hòabìnhians: the Önge and the Jehai (Peninsular Malaysia), along with groups from India and Papua New Guinea.
We also find a distinctive relationship between the group 1 samples and the Ikawazu Jōmon of Japan (IK002). Outgroup f3 statistics (11, 16) show that group 1 shares the most genetic drift with all ancient mainland samples and Jōmon (fig. S12 and table S4). All other ancient genomes share more drift with present-day East Asian and Southeast Asian populations than with Jōmon (figs. S13 to S19 and tables S4 to S11). This is apparent in the fastNGSadmix analysis when assuming six ancestral components (K = 6) (fig. S11), where the Jōmon sample contains East Asian components and components found in group 1. To detect populations with genetic affinities to Jōmon, relative to present-day Japanese, we computed D statistics of the form D(Japanese, Jōmon; X, Mbuti), setting X to be different presentday and ancient Southeast Asian individuals (table S22). The strongest signal is seen when X=Ma911 and La368 (group 1 individuals), showing a marginally nonsignificant affinity to Jōmon (11). This signal is not observed with X = Papuans or Önge, suggesting that the Jōmon and Hòabìnhians may share group 1 ancestry (11).
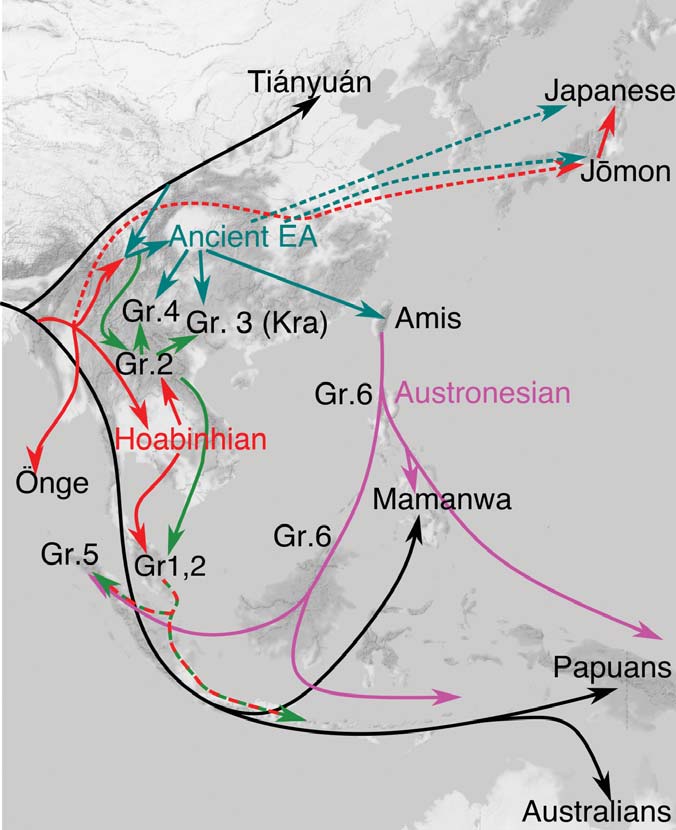
(…) Finally, the Jōmon individual is best-modeled as a mix between a population related to group 1/Önge and a population related to East Asians (Amis), whereas present-day Japanese can be modeled as a mixture of Jōmon and an additional East Asian component (Fig. 3 and fig. S29)
Interesting in relation to the oral communication of the SMBE O-03-OS02 Whole genome analysis of the Jomon remain reveals deep lineage of East Eurasian populations by Gakuuhari et al.:
Post late-Paleolithic hunter-gatherers lived throughout the Japanese archipelago, Jomonese, are thought to be a key to understanding the peopling history in East Asia. Here, we report a whole genome sequence (x1.85) of 2,500-year old female excavated from the Ikawazu shell-mound, unearthed typical remains of Jomon culture. The whole genome data places the Jomon as a lineage basal to contemporary and ancient populations of the eastern part of Eurasian continent, and supports the closest relationship with the modern Hokkaido Ainu. The results of ADMIXTURE show the Jomon ancestry is prevalent in present-day Nivkh, Ulchi, and people in the main-island Japan. By including the Jomon genome into phylogenetic trees, ancient lineages of the Kusunda and the Sherpa/Tibetan, early splitting from the rest of East Asian populations, is emerged. Thus, the Jomon genome gives a new insight in East Asian expansion. The Ikawazu shell-mound site locates on 34,38,43 north latitude, and 137,8, 52 east longitude in the central main-island of the Japanese archipelago, corresponding to a warm and humid monsoon region, which has been thought to be almost impossible to maintain sufficient ancient DNA for genome analysis. Our achievement opens up new possibilities for such geographical regions.
Related
- Model for the spread of Transeurasian (Macro-Altaic) communities with farming
- Genomics reveals four prehistoric migration waves into South-East Asia
- Population turnover in Remote Oceania shortly after initial settlement
- Language continuity despite population replacement in Remote Oceania
- Genomic history of South-East Asia: eastern Polynesians, Peninsular Malaysia and North Borneo
- Islands across the Indonesian archipelago show complex patterns of admixture
- Two more studies on the genetic history of East Asia: Han Chinese and Thailand
- Reconstructing the demographic history of the Himalayan and adjoining populations
- Ancient Di-Qiang people show early links with Han Chinese