Two recent interesting genetic papers:
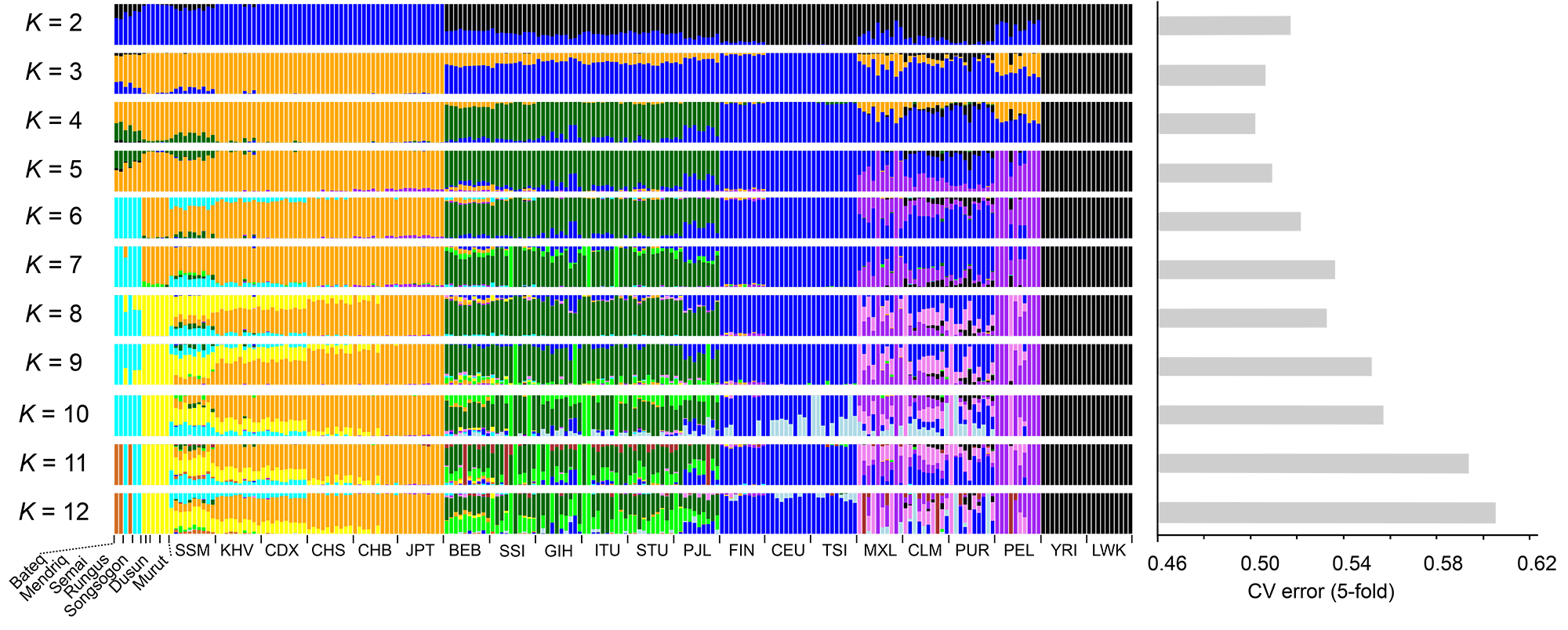
1. Open Access Investigating the origins of eastern Polynesians using genome-wide data from the Leeward Society Isles, by Hudjashov et al., at Scientific Reports (2018)
Abstract:
The debate concerning the origin of the Polynesian speaking peoples has been recently reinvigorated by genetic evidence for secondary migrations to western Polynesia from the New Guinea region during the 2nd millennium BP. Using genome-wide autosomal data from the Leeward Society Islands, the ancient cultural hub of eastern Polynesia, we find that the inhabitants’ genomes also demonstrate evidence of this episode of admixture, dating to 1,700–1,200 BP. This supports a late settlement chronology for eastern Polynesia, commencing ~1,000 BP, after the internal differentiation of Polynesian society. More than 70% of the autosomal ancestry of Leeward Society Islanders derives from Island Southeast Asia with the lowland populations of the Philippines as the single largest potential source. These long-distance migrants into Polynesia experienced additional admixture with northern Melanesians prior to the secondary migrations of the 2nd millennium BP. Moreover, the genetic diversity of mtDNA and Y chromosome lineages in the Leeward Society Islands is consistent with linguistic evidence for settlement of eastern Polynesia proceeding from the central northern Polynesian outliers in the Solomon Islands. These results stress the complex demographic history of the Leeward Society Islands and challenge phylogenetic models of cultural evolution predicated on eastern Polynesia being settled from Samoa.
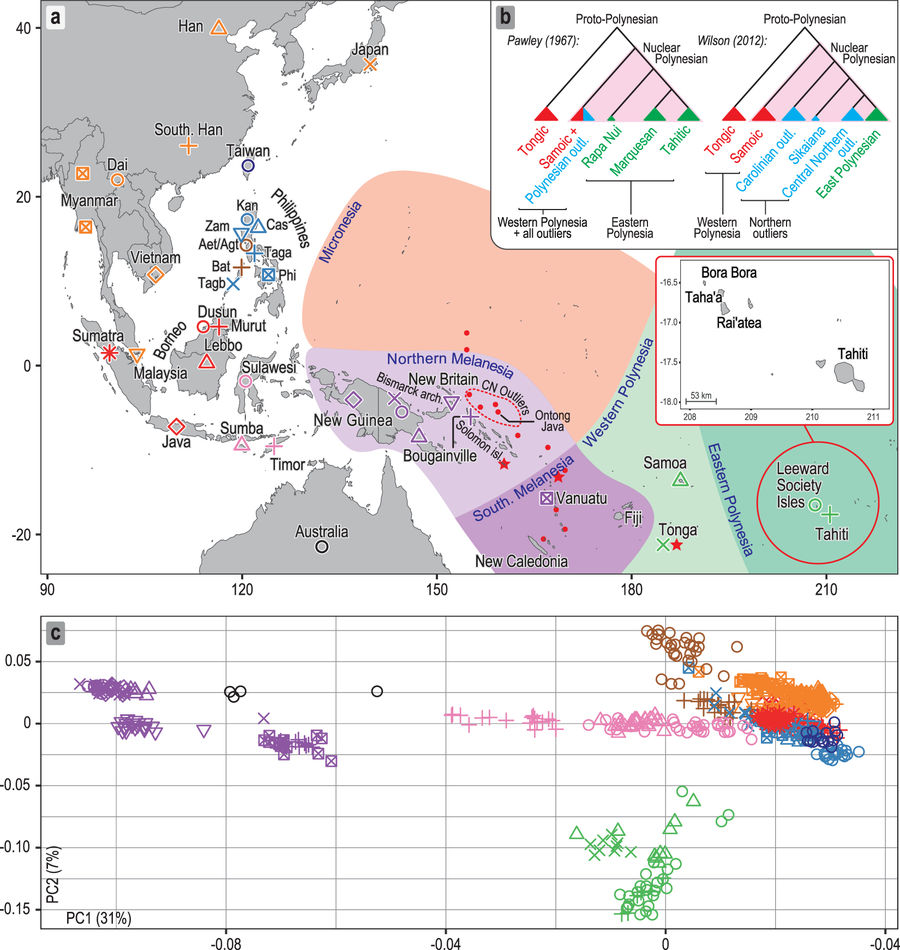
2. Genomic structure of the native inhabitants of Peninsular Malaysia and North Borneo suggests complex human population history in Southeast Asia, by Yew et al. at at Human Genetics (2018)
Abstract:
Southeast Asia (SEA) is enriched with a complex history of peopling. Malaysia, which is located at the crossroads of SEA, has been recognized as one of the hubs for early human migration. To unravel the genomic complexity of the native inhabitants of Malaysia, we sequenced 12 samples from 3 indigenous populations from Peninsular Malaysia and 4 native populations from North Borneo to a high coverage of 28–37×. We showed that the Negritos from Peninsular Malaysia shared a common ancestor with the East Asians, but exhibited some level of gene flow from South Asia, while the North Borneo populations exhibited closer genetic affinity towards East Asians than the Malays. The analysis of time of divergence suggested that ancestors of Negrito were the earliest settlers in the Malay Peninsula, whom first separated from the Papuans ~ 50–33 thousand years ago (kya), followed by East Asian (~ 40–15 kya), while the divergence time frame between North Borneo and East Asia populations predates the Austronesian expansion period implies a possible pre-Neolithic colonization. Substantial Neanderthal ancestry was confirmed in our genomes, as was observed in other East Asians. However, no significant difference was observed, in terms of the proportion of Denisovan gene flow into these native inhabitants from Malaysia. Judging from the similar amount of introgression in the Southeast Asians and East Asians, our findings suggest that the Denisovan gene flow may have occurred before the divergence of these populations and that the shared similarities are likely an ancestral component.
See also:
- Islands across the Indonesian archipelago show complex patterns of admixture
- Two more studies on the genetic history of East Asia: Han Chinese and Thailand
- Earliest modern humans outside Africa and ancient genomic history
- Genomic history of Northern Eurasians includes East-West and North-South gradients
- Review article about Ancient Genomics, by Pontus Skoglund and Iain Mathieson
- Review article on the origin of modern humans: the multiple-dispersal model and Late Pleistocene Asia
- Reconstructing the demographic history of the Himalayan and adjoining populations
- Ancient Di-Qiang people show early links with Han Chinese
- Indo-European and Central Asian admixture in Indian population, dependent on ethnolinguistic and geodemographic divisions
- Optimal Migration Routes of Initial Upper Palaeolithic Populations to Eurasia
- Genetic landscapes showing human genetic diversity aligning with geography