Looking for information on Novosvobodnaya samples from Wang et al. (2018) for my latest post, I stumbled upon this from the Supplementary Data 2 (download the Excel table):
Latvia_MN1.SG (ZVEJ26)
Skeletal element: petrous
Sample: Latvia_MN_dup.I4627.SG
Date: 4251-3976 calBCE
Location: Zvejnieki
mtDNA: U4a1
Y-DNA: R1b1a1a2
Coverage: 0.15
SNPs hit on autosomes: 167445
The data on Mathieson et al. (2018) is as follows:
I4627 (ZVEJ26)
Skeletal element: petrous
Origin: ThisStudy (New data; Individual first published in JonesNatureCommunications2017)
Sample: Latvia_MN
Date:4251-3976 calBCE (5280±55 BP, Ua-3639)
Location:Zvejnieki
mtDNA: U4a1
Y-DNA: R1b1a1a(xR1b1a1a2)
Coverage: 1.77
SNPs hit on autosomes: 686273Y-Chromosome derived SNPs: R1b1a1a:PF6475:17986687C->A; R1b1a1a:CTS3876:15239181G->C; R1b1a1a:CTS5577:16376495A->C; R1b1a1a:CTS9018:18617596C->T; R1b1a1a:FGC57:7759944G->A; R1b1a1a:L502:19020340G->C; R1b1a1a:PF6463:16183412C->A; R1b1a1a:PF6524:23452965T->C; R1b1a:A702:10038192G->A; R1b1a:FGC35:18407611C->T; R1b1a:FGC36:13822833G->T; R1b1a:L754:22889018G->A; R1b1a:L1345:21558298G->T; R1b1a:PF6249:8214827C->T; R1b1a:PF6263:21159055C->A; R1b1:CTS2134:14193384G->A; R1b1:CTS2229:14226692T->A; R1b1:L506:21995972T->A; R1b1:L822:7960019G->A; R1b1:L1349:22722580T->C; R1b:M343:2887824C->A; R1:CTS2565:14366723C->T; R1:CTS3123:14674176A->C; R1:CTS3321:14829196C->T; R1:CTS5611:16394489T->G; R1:L875:16742224A->G; R1:P238:7771131G->A; R1:P286:17716251C->T; R1:P294:7570822G->C; R:CTS207:2810583A->G; R:CTS2913:14561760A->G; R:CTS3622:15078469C->G; R:CTS7876:17722802G->A; R:CTS8311:17930099C->A; R:F33:6701239G->A; R:F63:7177189G->A; R:F82:7548900G->A; R:F154:8558505T->C; R:F370:16856357T->C; R:F459:18017528G->T; R:F652:23631629C->A; R:FGC1168:15667208G->C; R:L1225:22733758C->G; R:L1347:22818334C->T; R:M613:7133986G->C; R:M734:18066156C->T; R:P224:17285993C->T; R:P227:21409706G->C
Context of Latvia_MN1
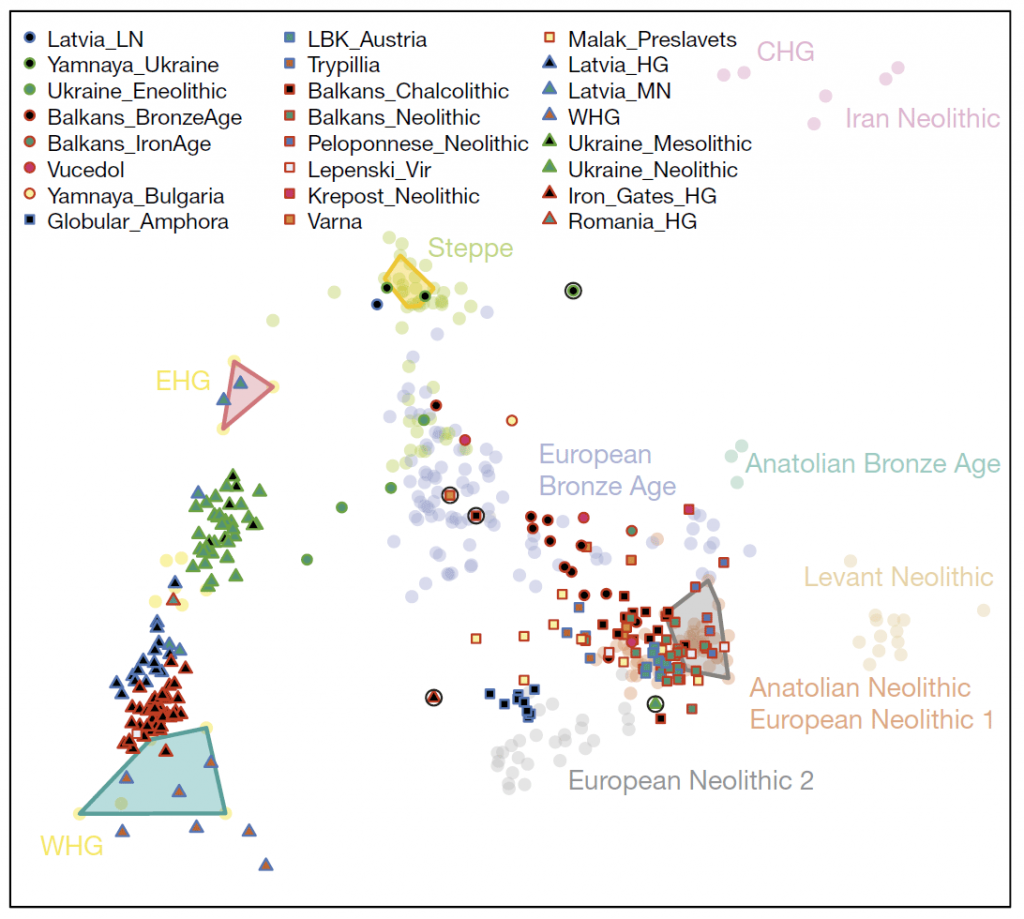
The Middle Neolithic is known to mark the westward expansion of Comb Ware and related cultures in North-Eastern Europe.
Mathieson et al. (2017 and 2018) had this to say about the Middle Neolithic in the Baltic:
At Zvejnieki in Latvia, using 17 newly reported individuals and additional data for 5 previously reported34 individuals, we observe a transition in hunter-gatherer-related ancestry that is opposite to that seen in Ukraine. We find that Mesolithic and Early Neolithic individuals (labelled ‘Latvia_HG’) associated with the Kunda and Narva cultures have ancestry that is intermediate between WHG (approximately 70%) and EHG (approximately 30%), consistent with previous reports34–36(Supplementary Table 3). We also detect a shift in ancestry between Early Neolithic individuals and those associated with the Middle Neolithic Comb Ware complex (labelled ‘Latvia_MN’), who have more EHG-related ancestry; we estimate that the ancestry of Latvia_MN individuals comprises 65% EHG-related ancestry, but two of the four individuals appear to be 100% EHG in principal component space (Fig. 1b).
Other samples and errors on Y-SNP calls
The truth is, this is another sample (Latvia_MN_dup.I4627.SG) from the same individual ZVEJ26.
There is another sample used for the analysis of ZVEJ26, with the same data as in Mathieson et al. (2018), i.e. better coverage, and Y-DNA R1b1a1a(xR1b1a1a2).
Most samples in the tables from Wang et al. (2018) seem to be classified correctly, as in previous papers, but for:
- Blätterhöhle Cave sample from Lipson et al. (2017), wrongly classified (again) as R1b1a1a2a1a2a1b2 (I am surprised no R1b-autochtonous-continuity-fan rushed to proclaim something based on this);
- Mal’ta 1 sample from Raghavan et al. (2013) as R1b1a1a2;
- Iron Gates HG, Schela Cladovey from Gonzalez Fortes (2017) as R1b1a1a2;
- Oase1 from Fu (2015) as N1c1a;
- samples from Skoglund et al. (2017) from Africa also wrongly classified as R1b1a1a2 and subclades.
It seems therefore that the poor coverage / SNPs hit on autosomes is the key common factor here for these Y-SNP calls, and so it is in the Zvejnieki MN1 duplicated sample. Anyway, if all Y-SNP calls come from the same software applied to all data, and this is going to be used in future papers, this seems to be a great improvement compared to Narasimhan et al. (2018)…
EDIT (25 JUN 2018): I have been reviewing some more papers apart from Mathieson et al. (2018) and Olalde et al. (2018) to compare the reported haplogroups, and there seems to be many potential errors (or updated data, difficult to say sometimes, especially when the newly reported haplogroup is just one or two subclades below the reported one in ‘old’ papers), not only those listed above.
The sample accession number in the European Nucleotide Archive (ENA) is SAMEA45565168 (Latvia_MN1/ZVEJ26) (see here), in case anyone used to this kind of analysis wishes to repeat the Y-SNP calls on both samples.
EDIT (25 JUN 2018): Added that it is another sample with lesser coverage from the same ZVEJ26 individual.
Related:
- The unique elite Khvalynsk male from a Yekaterinovskiy Cape burial
- Haplogroup R1b-L51 in Khvalynsk samples from the Samara region dated ca. 4250-4000 BC
- The Indo-European demic diffusion model, and the “R1b – Indo-European” association
- Early Indo-Iranian formed mainly by R1b-Z2103 and R1a-Z93, Corded Ware out of Late PIE-speaking migrations
- Proto-Indo-European homeland south of the Caucasus?
- On the potential origin of Caucasus hunter-gatherer ancestry in Eneolithic steppe cultures