Open access New insights from Thailand into the maternal genetic history of Mainland Southeast Asia, by Kutanan et al. Eur. J. Hum. Genet. (2018) 26:898–911
Abstract (emphasis mine):
Tai-Kadai (TK) is one of the major language families in Mainland Southeast Asia (MSEA), with a concentration in the area of Thailand and Laos. Our previous study of 1234 mtDNA genome sequences supported a demic diffusion scenario in the spread of TK languages from southern China to Laos as well as northern and northeastern Thailand. Here we add an additional 560 mtDNA genomes from 22 groups, with a focus on the TK-speaking central Thai people and the Sino-Tibetan speaking Karen. We find extensive diversity, including 62 haplogroups not reported previously from this region. Demic diffusion is still a preferable scenario for central Thais, emphasizing the expansion of TK people through MSEA, although there is also some support for gene flow between central Thai and native Austroasiatic speaking Mon and Khmer. We also tested competing models concerning the genetic relationships of groups from the major MSEA languages, and found support for an ancestral relationship of TK and Austronesian-speaking groups.
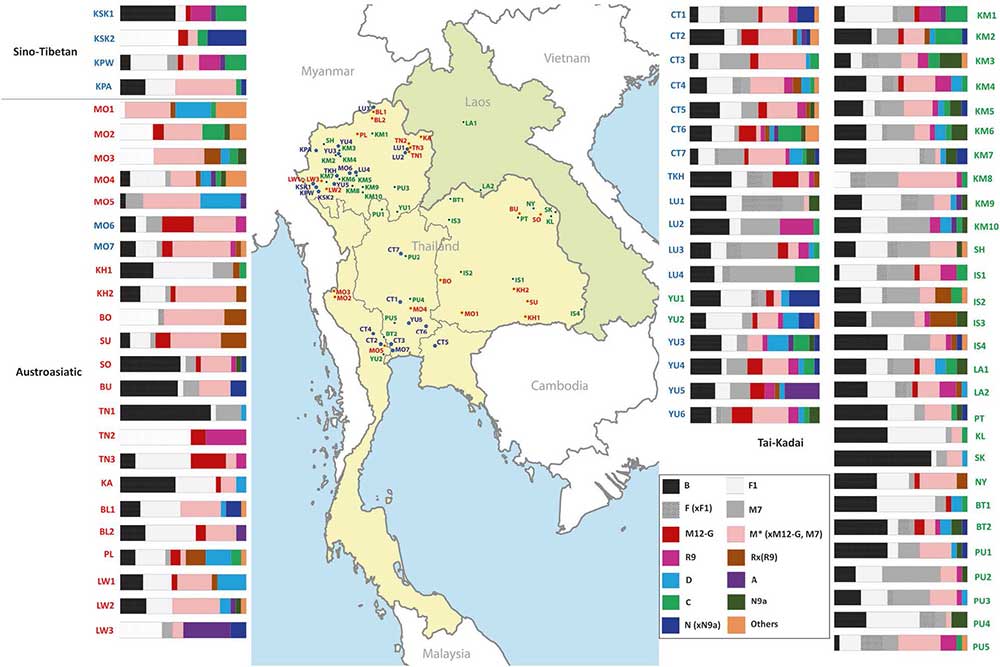
Interesting excerpts:
Finally, we used simulations to test hypotheses concerning the genetic relationships of groups belonging to different language families. We found that Starosta’s model [11] provided the best fit to the mtDNA data; however, Sagart’s model [9, 10] was also highly supported. These two models both postulate a close linguistic affinity between TK and AN. Although genetic relatedness between TK and AN groups has been previously studied [7, 46, 47], to our knowledge this is the first study to use demographic simulations to select the best-fitting model. Our results support the genetic relatedness of TK and AN groups, which might reflect a postulated shared ancestry among the proto-Austronesian populations of coastal East Asia [48].
Specifically, the best-fitting model suggests that after separation of the prehistoric TK from AN stocks around 5–6 kya in Southeast China, the TK spread southward throughout MSEA around 1–2 kya by a demic diffusion process, accompanied by population growth but with at most minor admixture with the autochthonous AA groups. Meanwhile, the prehistorical AN ancestors entered Taiwan and dispersed southward throughout ISEA, with these two expansions later meeting in western ISEA. The lack of mtDNA haplogroups associated with the expansion out of Taiwan in our Thai/Lao samples has two possible explanations: either the Out of Taiwan expansion did not reach MSEA (at least, in the area of present-day Thailand and Laos); or, if the prehistoric AN migrated through this area, their mtDNA lineages do not survive in modern Thai/Lao populations. Ancient DNA studies in MSEA would further clarify this issue. Moreover, although mtDNA analyses are informative in elucidating genetic perspectives in geographically and linguistically related populations, they have an obvious limitation in that they only provide insights into the maternal history of populations. Future studies of Y chromosomal and genome-wide data will provide further insights into the genetic history of Thai/Lao populations and the role of factors such as post-marital residence patterns and migration in shaping the genetic structure of the region.
Starosta’s chapter referred to in the paper is Proto-East Asian and the origin and dispersal of the languages of East and Southeast Asia and the Pacific.
Related:
- Ancient genomes document multiple waves of migration in south-east Asian prehistory
- Genomics reveals four prehistoric migration waves into South-East Asia
- Population turnover in Remote Oceania shortly after initial settlement
- Language continuity despite population replacement in Remote Oceania
- Genomic history of South-East Asia: eastern Polynesians, Peninsular Malaysia and North Borneo
- Islands across the Indonesian archipelago show complex patterns of admixture
- Two more studies on the genetic history of East Asia: Han Chinese and Thailand
- Reconstructing the demographic history of the Himalayan and adjoining populations
- Ancient Di-Qiang people show early links with Han Chinese