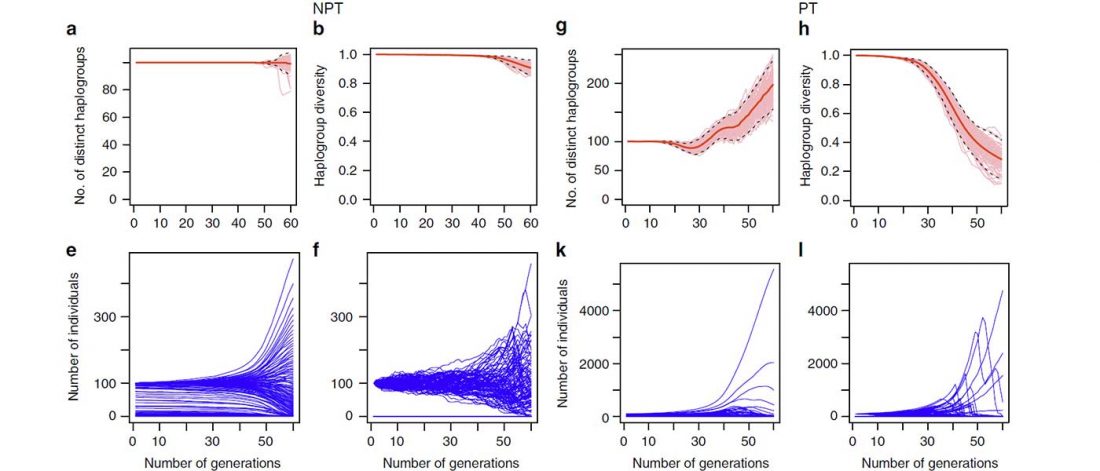
Open access study Cultural hitchhiking and competition between patrilineal kin groups explain the post-Neolithic Y-chromosome bottleneck, by Zeng, Aw, and Feldman, Nature Communications (2018).
Abstract (emphasis mine):
… Read the rest “Post-Neolithic Y-chromosome bottleneck explained by cultural hitchhiking and competition between patrilineal clans”In human populations, changes in genetic variation are driven not only by genetic processes, but can also arise from cultural or social changes. An abrupt population bottleneck specific to human males has been inferred across several Old World (Africa, Europe, Asia) populations 5000–7000 BP. Here, bringing together anthropological theory, recent population genomic studies and mathematical models, we propose a sociocultural hypothesis, involving the formation of patrilineal kin groups and intergroup competition among