Informal report by Bulgarian archaeologist Svetoslav Stamov in 7/8 TV, from data collected by the Reich Lab for their future paper on South-Eastern Europe.
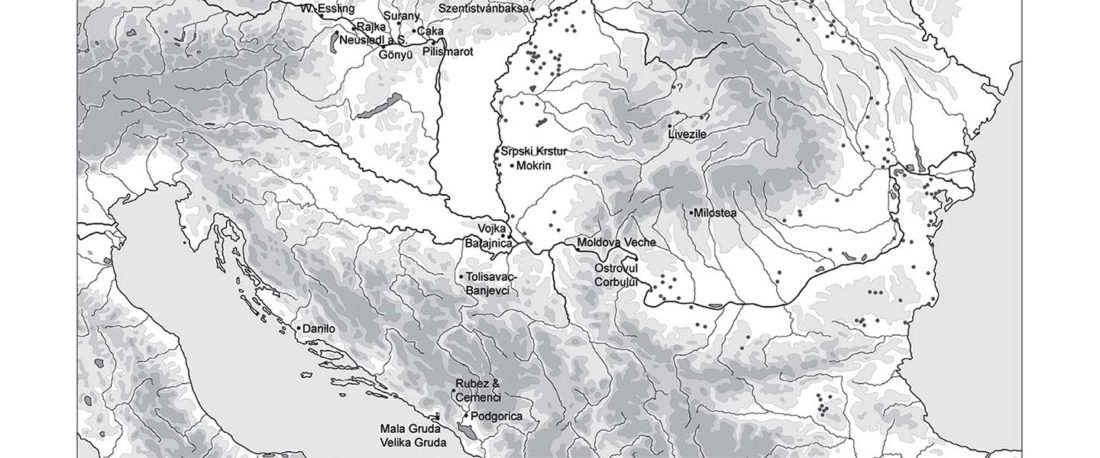
As can be seen from the TV captions below, this is the earliest R1b-P310 from Yamnaya or Yamnaya-related individuals in Early Bronze Age contexts from Bulgaria. In fact, its appearance together with a R1b-Z2103 lineage (and another undefined R1b-M269) shows once again that the earliest R1b-L23 bottlenecks were associated with Proto-Indo-Europeans.
Lacking a precise periodization, location, or proper cultural context in the spreadsheet, it is impossible to know whether they belong to Khvalynsk-related cultures … Read the rest “West Yamnaya settlers like Early Bell Beakers: R1b-P310 and R1b-Z2103”