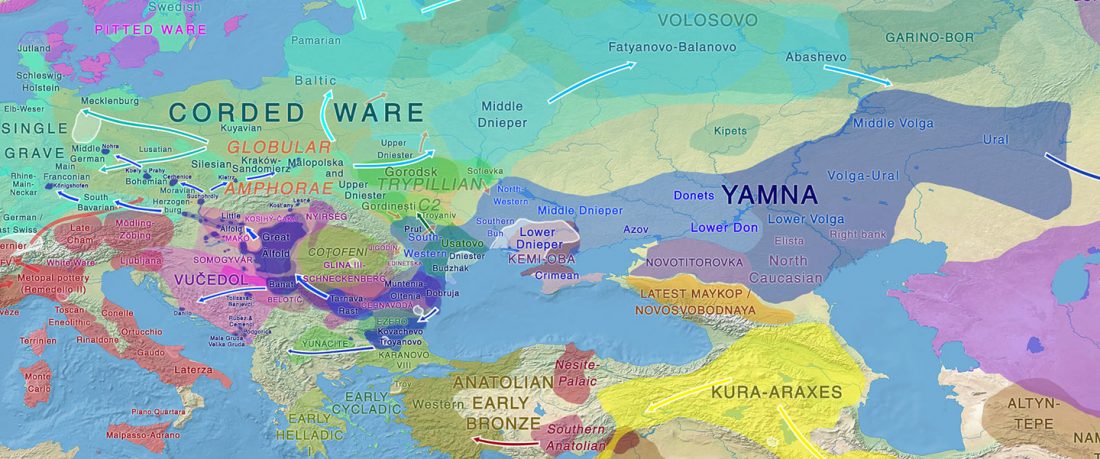
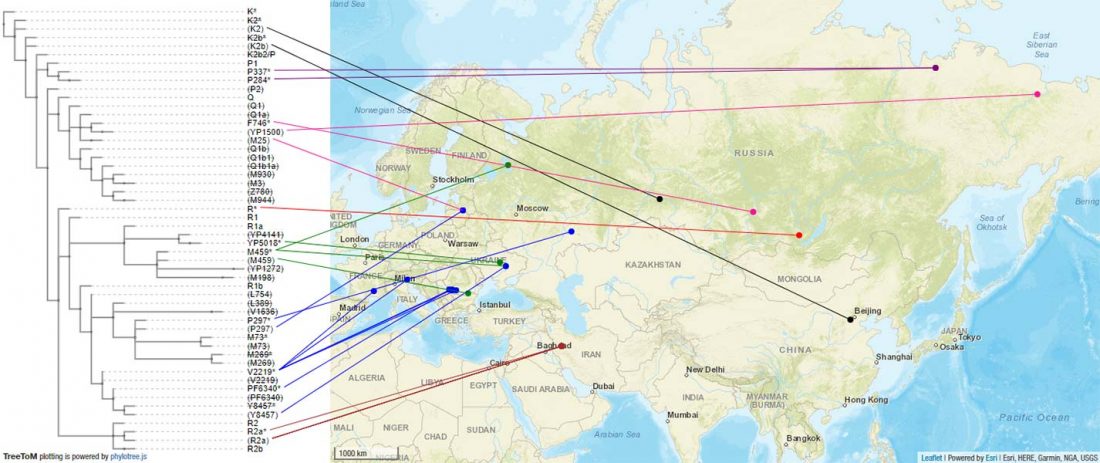
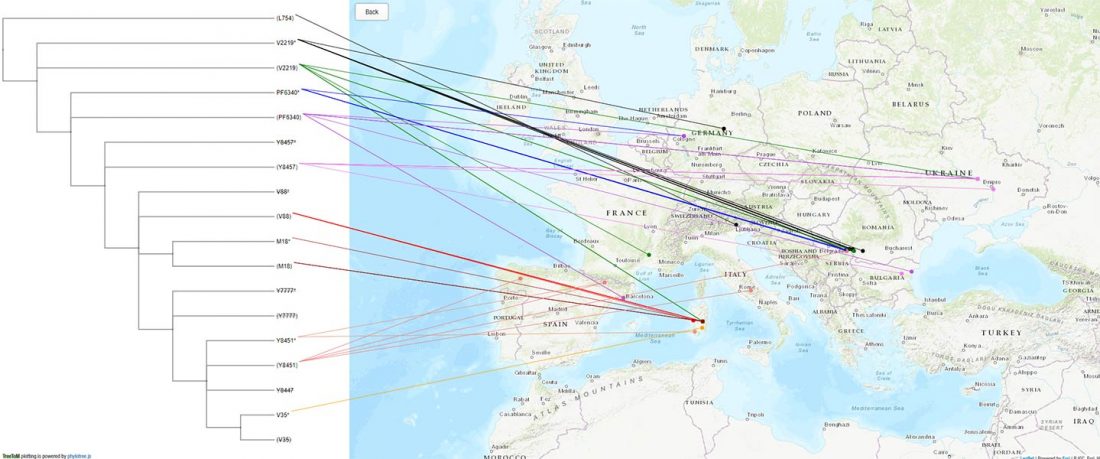
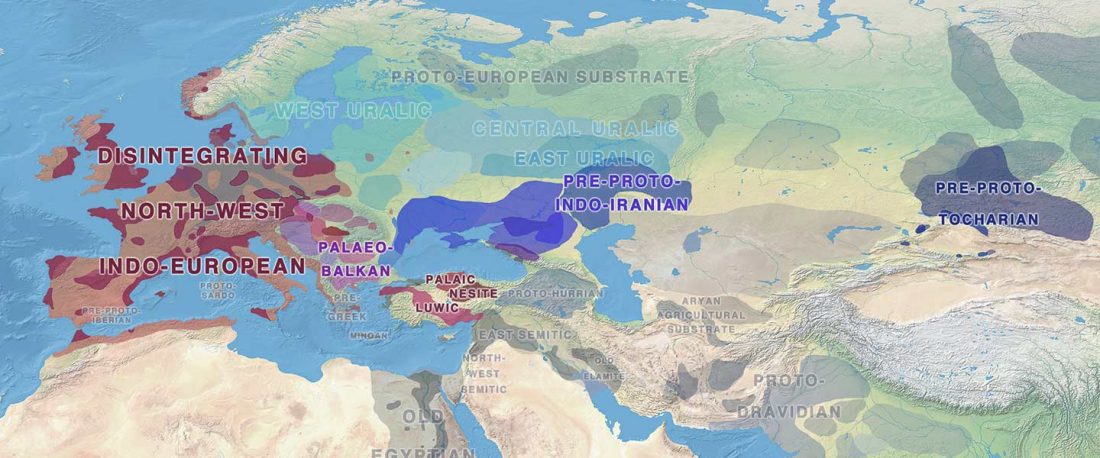
Another interesting finding from Human auditory ossicles as an alternative optimal source of ancient DNA, by Sirak et al. (2020):
A sample classified as Italy Middle Bronze Age from Olmo di Nogara (ca. 1400-1200 BC), who is R1b-L51 (xP311, xL52, xL151), CTS6889+ (T->C, 1 read). See YFull’s corresponding R-S1161.
This sample probably belongs to individual 309 (35-45 yo), and the female sampled to 323 (30-40 yo), both referenced as from the following study:
… Read the rest “Italo-Venetic peoples related patrilineally to Terramare elites”Canci A, Contursi D, Fornaciari G. 2005. La necropoli dell’età del bronzo di Olmo di Nogara (Verona): primi risultati