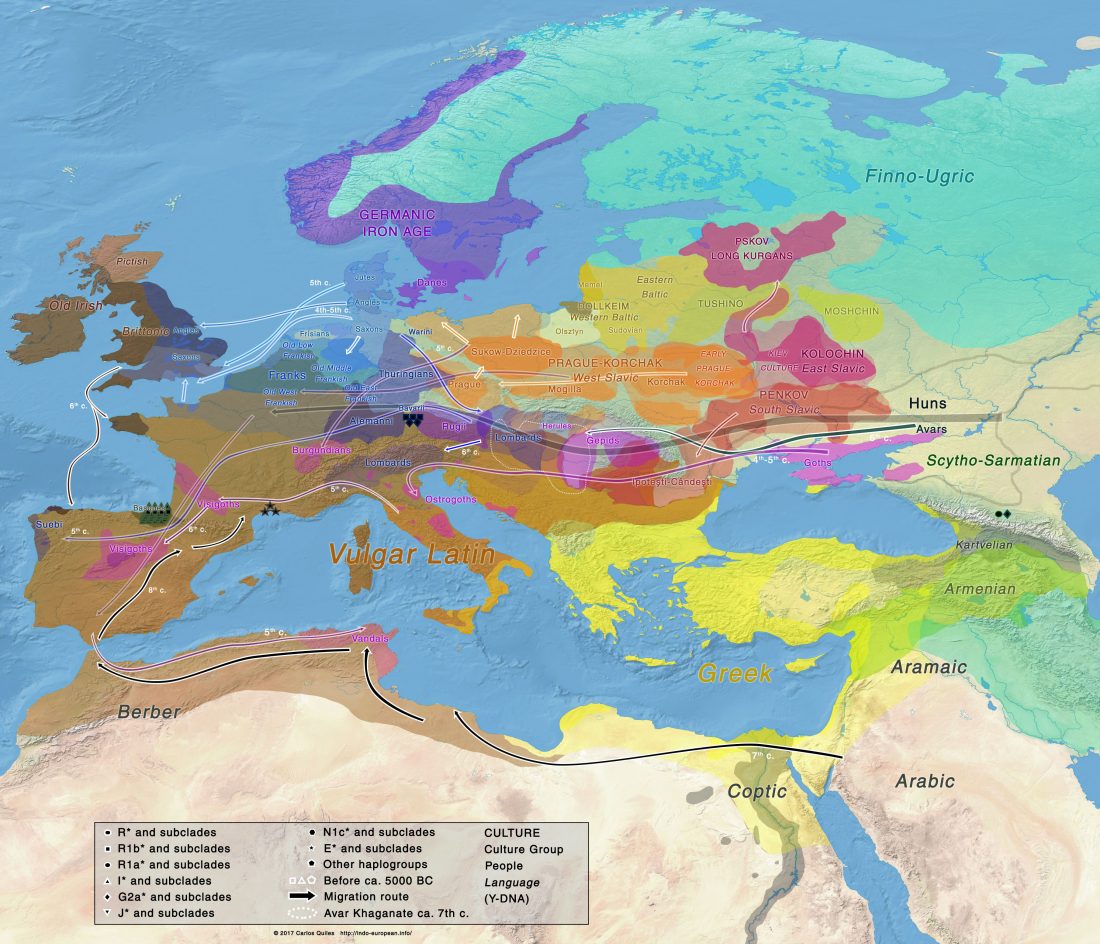
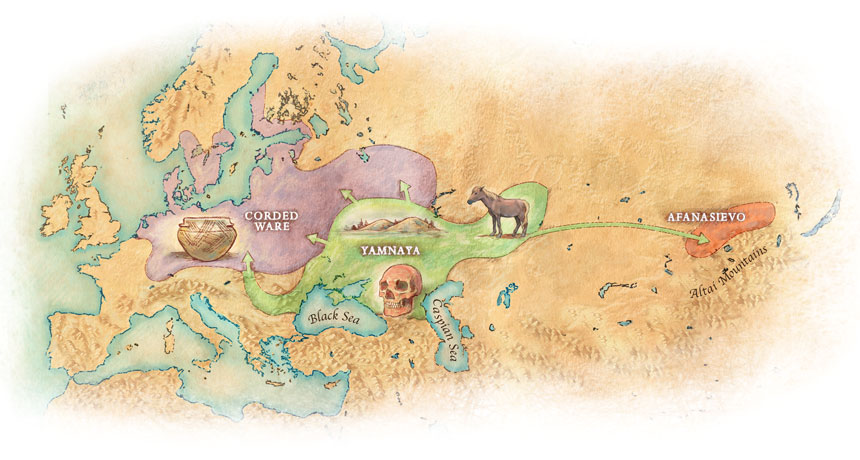
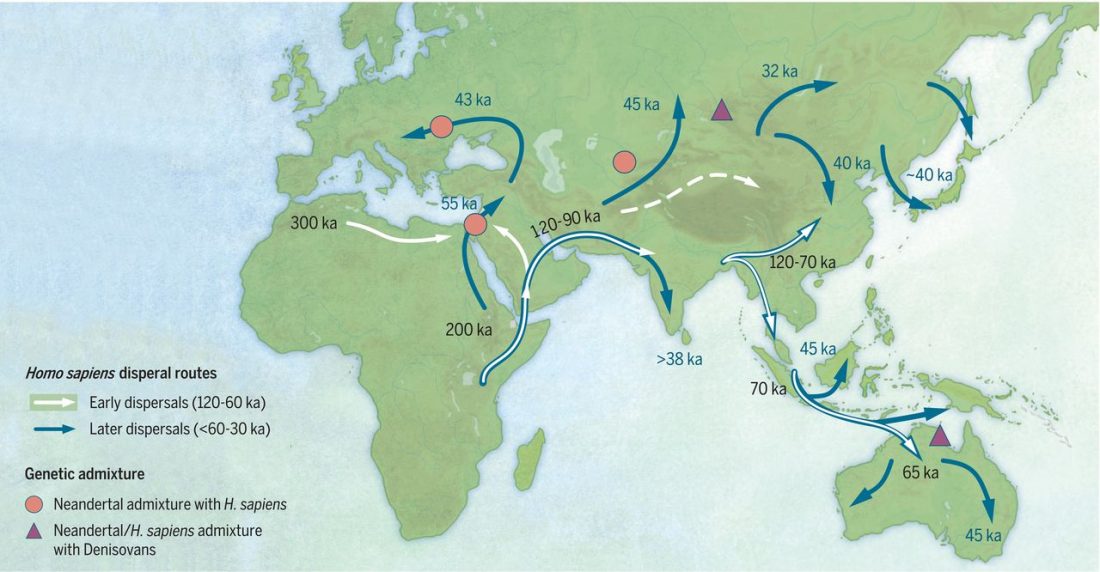
Open access Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans, by Krause-Kyora et al., Nature Communications (2018)
Abstract:
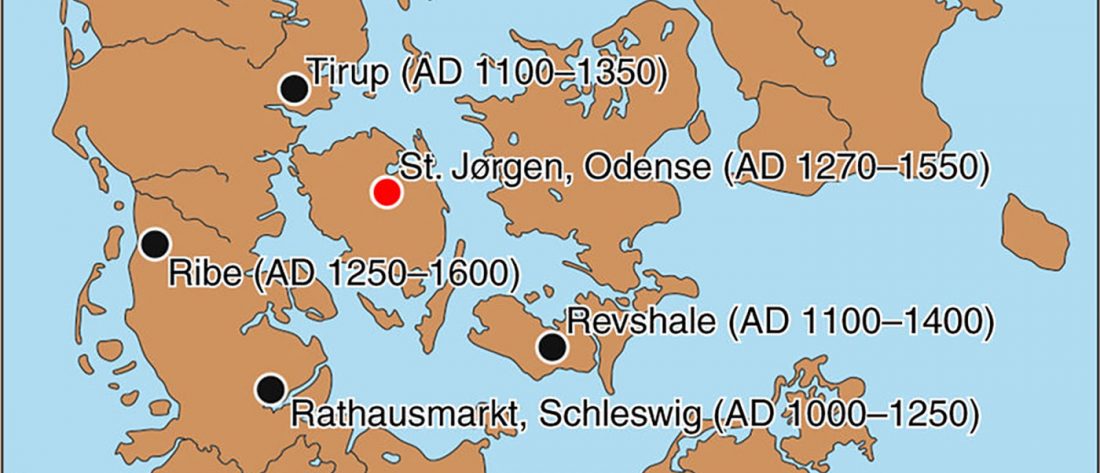
… Read the rest “Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans”Leprosy, a chronic infectious disease caused by Mycobacterium leprae (M. leprae), was very common in Europe till the 16th century. Here, we perform an ancient DNA study on medieval skeletons from Denmark that show lesions specific for lepromatous leprosy (LL). First, we test the remains for M. leprae DNA to confirm the infection status of the individuals and to assess the bacterial diversity. We assemble 10 complete M. leprae genomes that all differ from each other. Second,