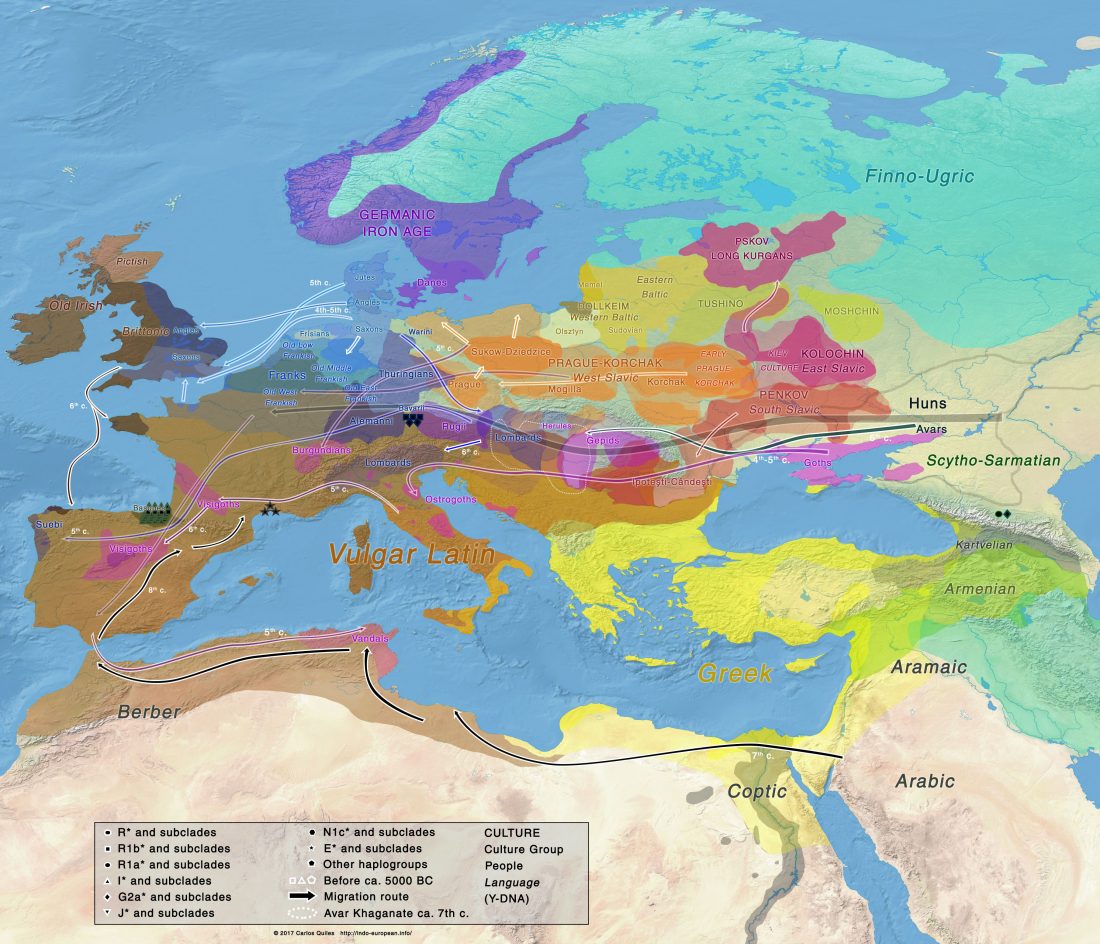
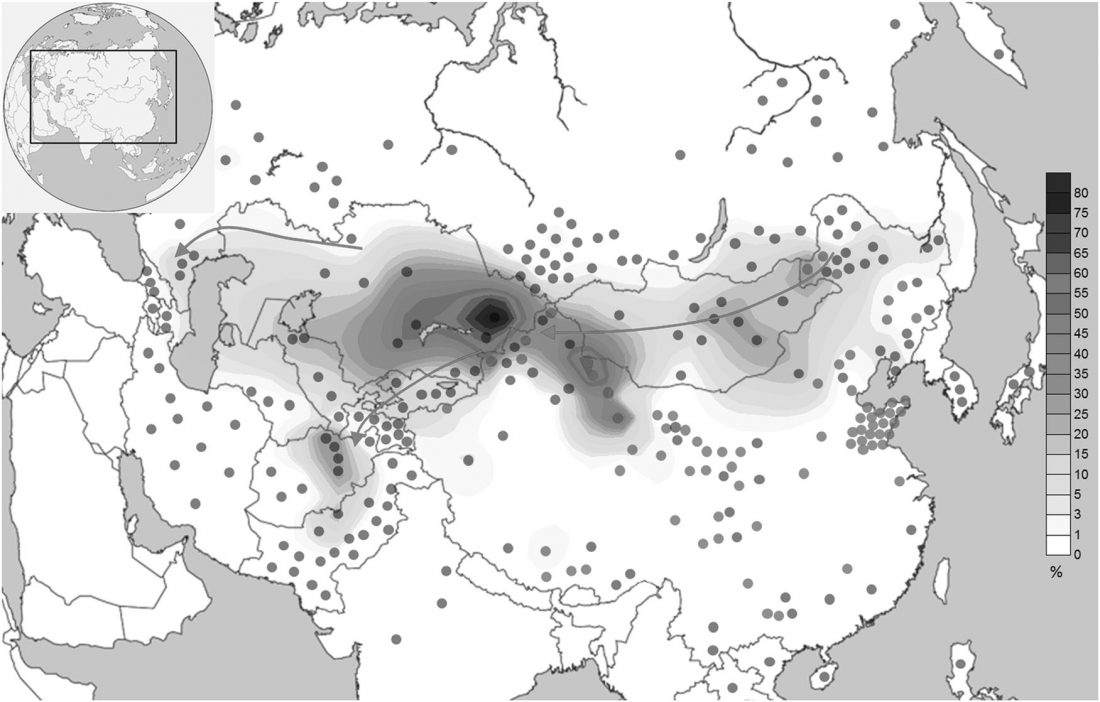
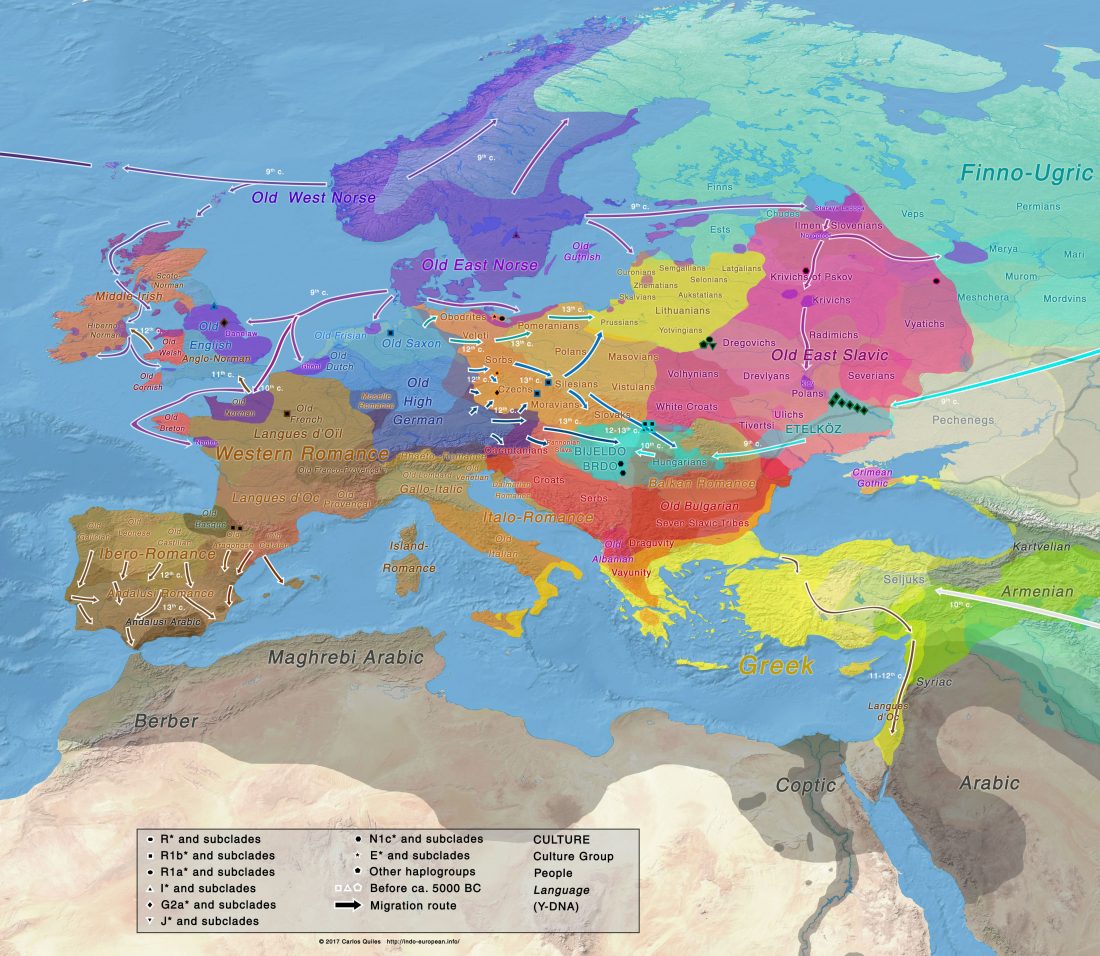
Y-DNA relevant in the postgenomic era, mtDNA study of Iron Age Italic population, and reconstructing the genetic history of Italians
Open Access Annals of Human Biology (2018), Volume 45, Issue 1, with the title Human population genetics of the Mediterranean.
Among the most interesting articles (emphasis mine):
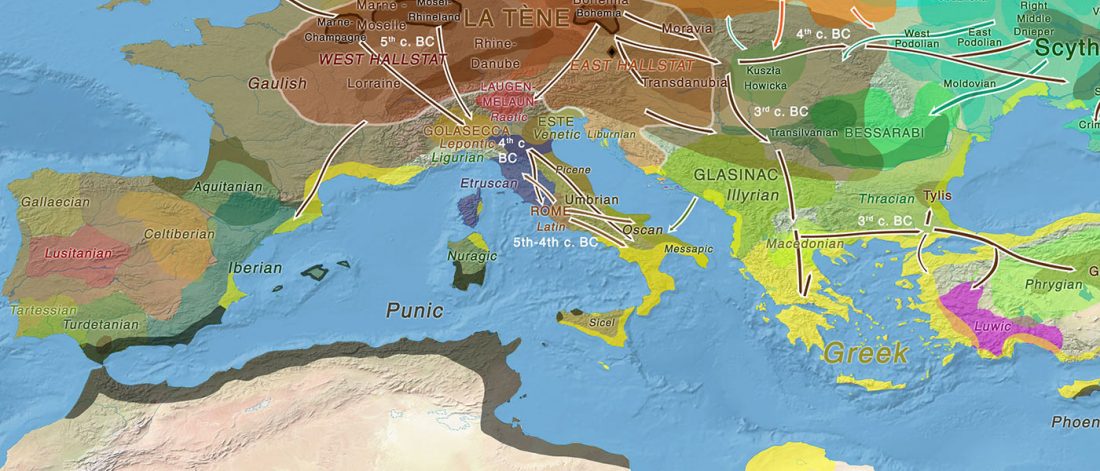
Iron Age Italic population genetics: the Piceni from Novilara (8th–7th century BC), by Serventi, Panicucci, Bodega, et al.
… Read the rest “Y-DNA relevant in the postgenomic era, mtDNA study of Iron Age Italic population, and reconstructing the genetic history of Italians”Background: Archaeological data provide evidence that Italy, during the Iron Age, witnessed the appearance of the first communities with well defined cultural identities. To date, only a few studies report genetic data about these populations and, in particular, the Piceni have never been analysed.
Aims: To provide new data about mitochondrial DNA (mtDNA)