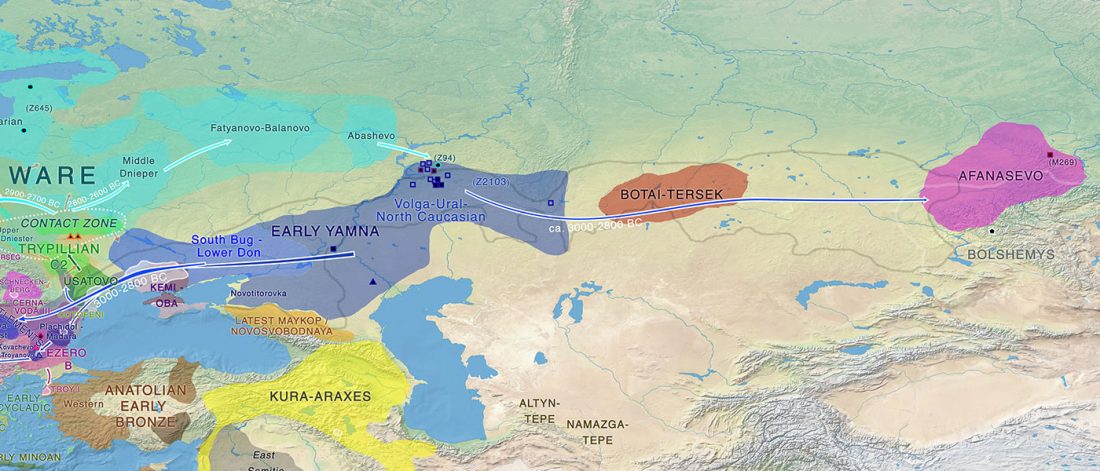
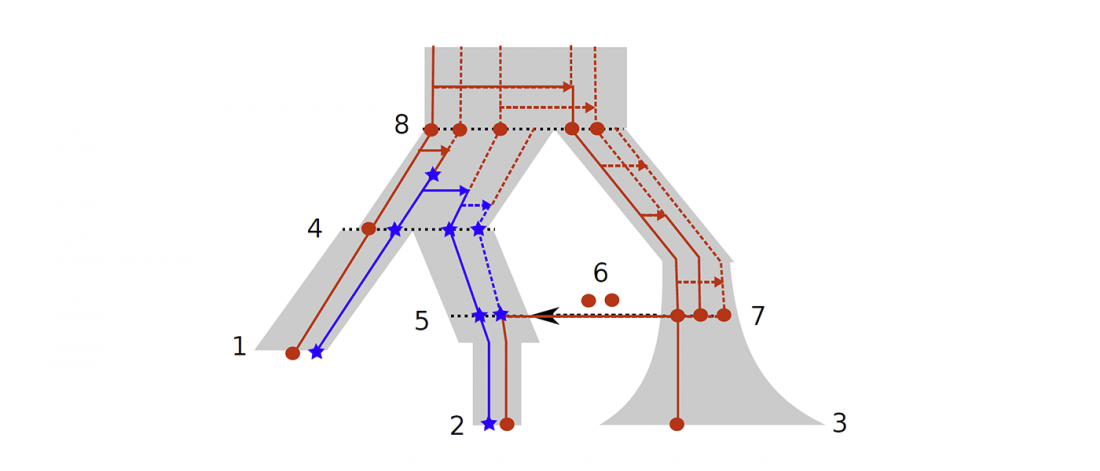
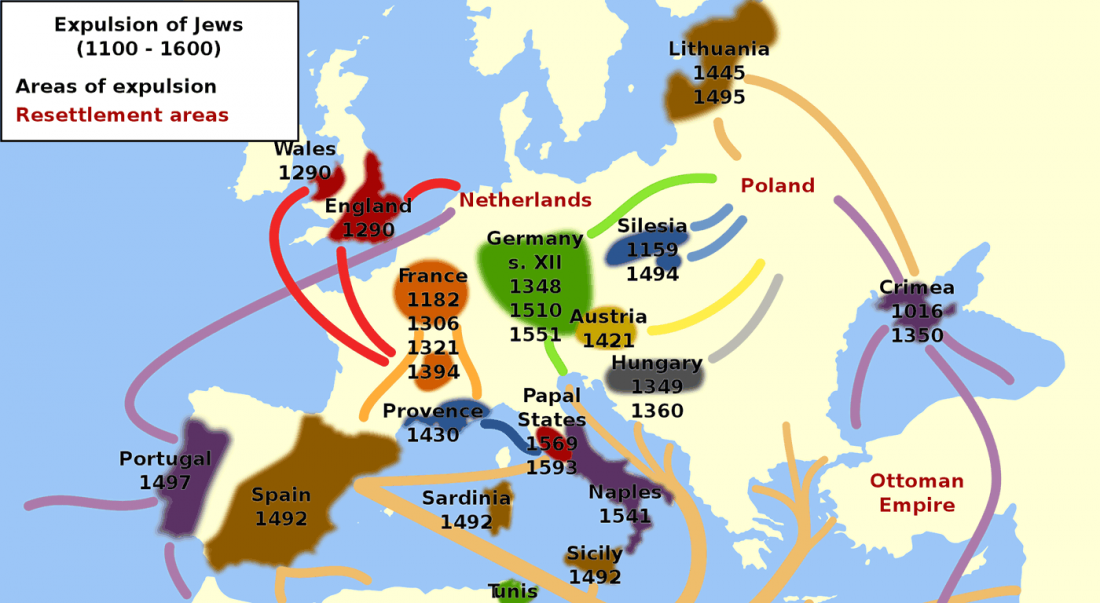
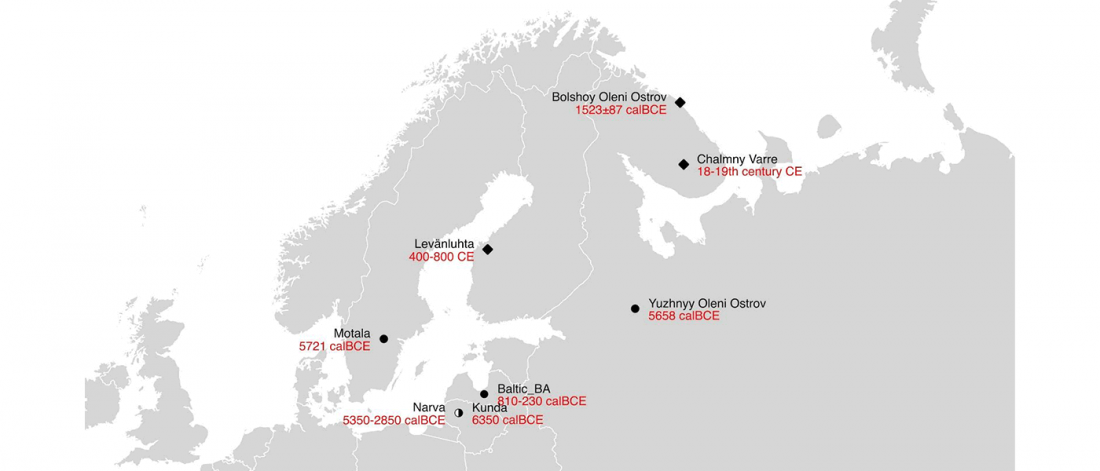
Distribution of Southern Iberian haplogroup H indicates exchanges in the western Mediterranean
Recent open access paper The distribution of mitochondrial DNA haplogroup H in southern Iberia indicates ancient human genetic exchanges along the western edge of the Mediterranean, by Hernández, Dugoujon, Novelletto, Rodríguez, Cuesta and Calderón, BMC Genetics (2017).
Abstract (emphasis mine):
… Read the rest “Distribution of Southern Iberian haplogroup H indicates exchanges in the western Mediterranean”Background
The structure of haplogroup H reveals significant differences between the western and eastern edges of the Mediterranean, as well as between the northern and southern regions. Human populations along the westernmost Mediterranean coasts, which were settled by individuals from two continents separated by a relatively narrow body of water, show the highest frequencies of mitochondrial haplogroup H. These