Recent paper A dynastic elite in monumental Neolithic society, by Cassidy et al. Nature (2020) 582:384–388.
Interesting excerpts (emphasis mine):
Neolithic Admixture
We sampled remains from all of the major Irish Neolithic funerary traditions: court tombs, portal tombs, passage tombs, Linkardstown-type burials and natural sites. Within this dataset, the earliest Neolithic human remains from the island—interred at Poulnabrone portal tomb14—are of majority ‘Early_Farmer’ ancestry (as defined by ADMIXTURE modelling), and show no evidence of inbreeding, which implies that, from the very onset, agriculture was accompanied by large-scale maritime colonization. Our ADMIXTURE and ChromoPainter analyses do not distinguish between the Irish and British Neolithic populations. They also confirm previous reports that samples from the Early Neolithic of Spain are the best proxy source of their Early_Farmer ancestry, which emphasizes the importance of Atlantic and Mediterranean waterways in their forebearers’ expansions.
Overall, no increase in inbreeding is seen through time in Neolithic Ireland, which indicates that communities maintained sufficient size and communication to avoid matings between relatives of the fifth degree or closer (…)
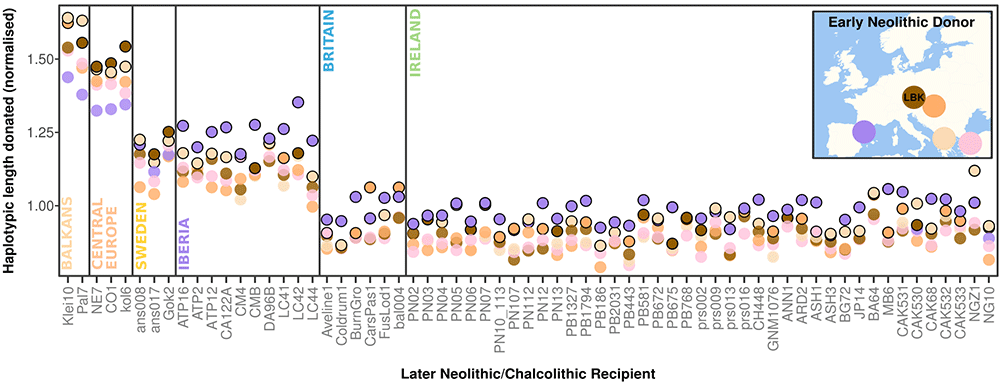
Kinship in prestigious structures
A second centre of the passage tomb tradition is found 150 km to the west of Newgrange, near the Atlantic coast. This centre comprises the mega-cemeteries of Carrowmore and Carrowkeel, which have origins that pre-date the construction of Newgrange by several centuries; depositions at Carrowkeel continued until at least the end of the Neolithic. Using analyses based on both single-nucleotide polymorphisms (SNPs) and haplotypes, we uncover a web of relatedness that connects these sites to both Newgrange and the atypical Millin Bay megalith on the northeast coast, which has previously been recognized as part of the broader passage tomb tradition on the basis of its artwork and morphological features.
(…)we find that the earliest passage tomb genome in the dataset (designated car004)—interred within the focal monument at Carrowmore— has detectable distant kinship with NG10 [the “god-king” fruit of first-order incest], as well as with other later individuals from Carrowkeel and Millin Bay (designated CAK533 and MB6, respectively). A similar kinship coefficient (≥6th degree) is also seen between NG10 and another individual from Carrowkeel (designated CAK532), demonstrating familial ties between several of the largest hubs of the tradition. (…)
Taken together, we favour the interpretation that the haplotypic structure within our dataset is driven by excessive identity-by-descent sharing between passage tomb samples, which implies non-random mating across large territories of the island. A high degree of social complexity would be required to achieve this (…).
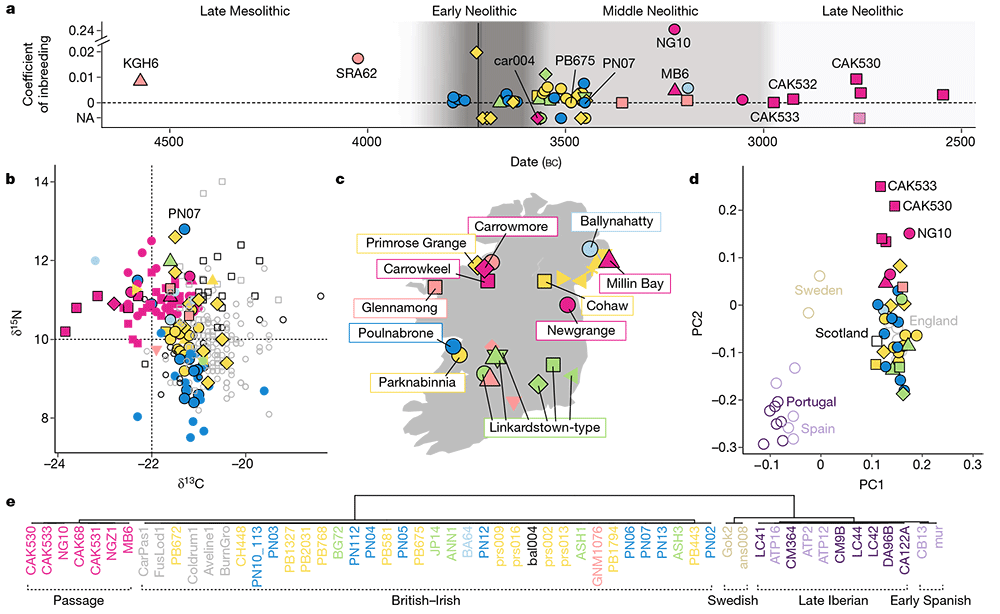
a, Timeline of analysed Irish genomes with inbreeding coefficients are shown for those with sufficient coverage. All dates are direct and calibrated, excluding that< for individual CAK534 (translucent). The key for the sample sites is given in c. Theearliest widespread evidence of Neolithic activity (house horizon) is marked with a black line. The Irish Neolithic ends at about 2500 bc. NA, not applicable.
b, Stable isotope values for samples from the Irish and British Neolithic (n = 292). The key for the Irish samples is given in c, and samples included in the ancient DNA analysis are outlined in black. British samples are shown as hollow shapes; black, Scotland; grey, England or Wales; circles, pre-3400 bc; squares, post-3400 bc. An infant with Down syndrome (PN07) is labelled; this individual showed isotope values consistent with a high trophic level.
c, Site locations for Irish individuals sampled or included in this study coloured by burial type: yellow, court tomb; blue, portal tomb; green, Linkardstown-type; magenta,passage tomb and related; light pink, natural sites; and light blue, the unclassified Ballynahatty7 megalith. Sites outlined in black were included in ancient DNA analysis.
d, ChromoPainter13 principal component (PC) analysis of individuals from the Atlantic seaboard of majority Early_Farmer ancestry (n = 57), generated using a matrix of haplotypic length-sharing. Passage tomb outliers in Fig. 2d are labelled.
e, fineSTRUCTURE dendrogram derived from the same matrix as in d with five consistent clusters.
Kinship in other groups
The simpler court and portal tombs lack the artwork and prestigious grave goods of the passage tomb tradition, and are arguably a manifestation of smaller-scale, lineage-based societies. These architectures do not typically occur within cemeteries of passage tombs—although exceptions do exist, including a court tomb constructed beside Carrowmore with a reported instance of inter-site kinship. We find evidence of both distant kinship and societal structure between another pair of distinct, but neighbouring, megaliths (10 km apart)—the Poulnabrone portal tomb14 and the Parknabinnia court tomb27. Their sampled cohorts (the majority of which are males) show a significant difference in the frequency of two Y chromosome haplogroups (P = 0.035, Fisher’s exact test), as well as a dietary difference. Given that there is a lack of close kin within either tomb, we exclude small family groups as their sole proprietors and interpret our findings as the result of broader social differentiation with an emphasis on patrilineal descent. The double occurrence of a rare Y haplogroup (H2a) among the individualized male Linkardstown burials of the southeast of the island provides further evidence of the importance of patrilineal ancestry in these societies, as does the predominance of a single Y haplogroup (I-M284) across the Irish and British Neolithic population
It is interesting to notice yet again strict patrilineal descent among regional Megalithic culture groups in Ireland, similar to previously reported ones in Sánchez-Quinto et al. (2019) for Sweden or Great Britain. However, there is no clear ancestral connection to all of them, which – coupled with the different attested traditions – suggests that the expansion of megalithic tombs might have been the product of cultural diffusion, too. Or perhaps it spread with stepped demic diffusion events whose complex development is difficult to unravel with the current data and tools.
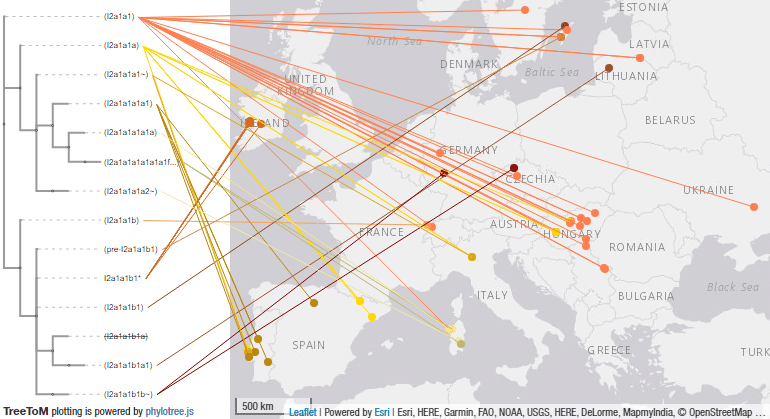
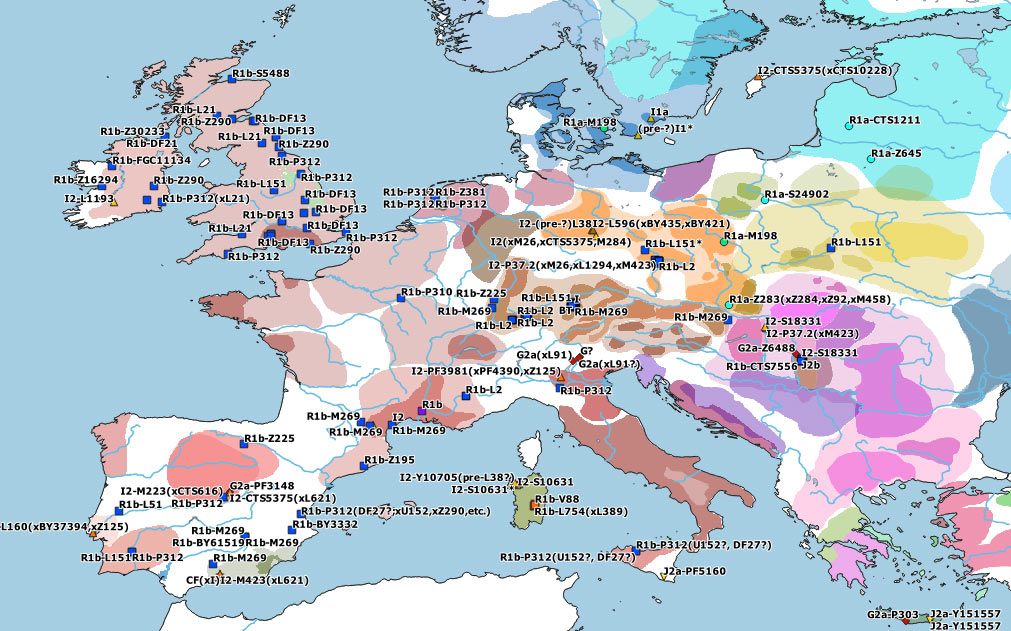
Neolithic expansion of I2 subclades
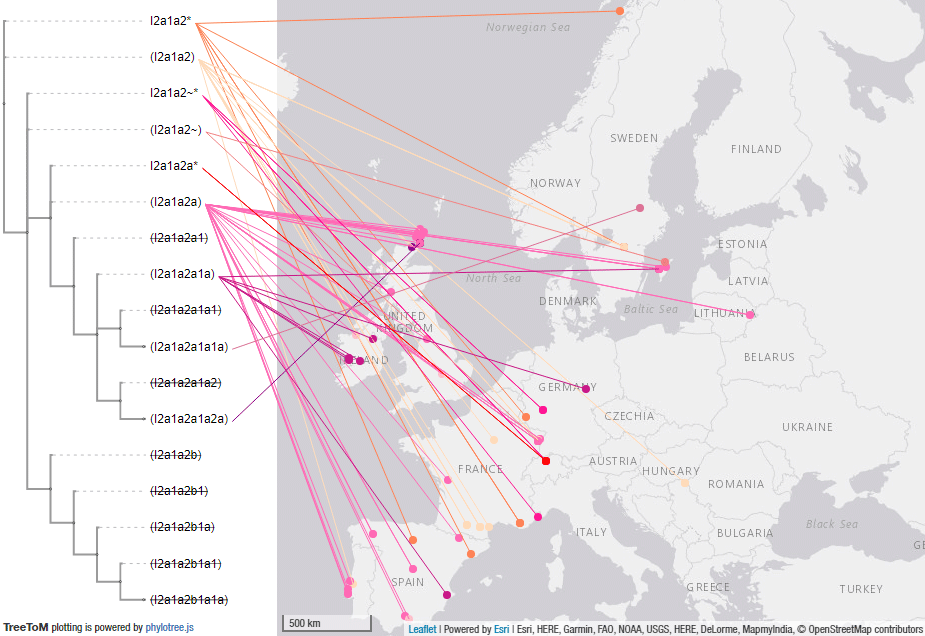
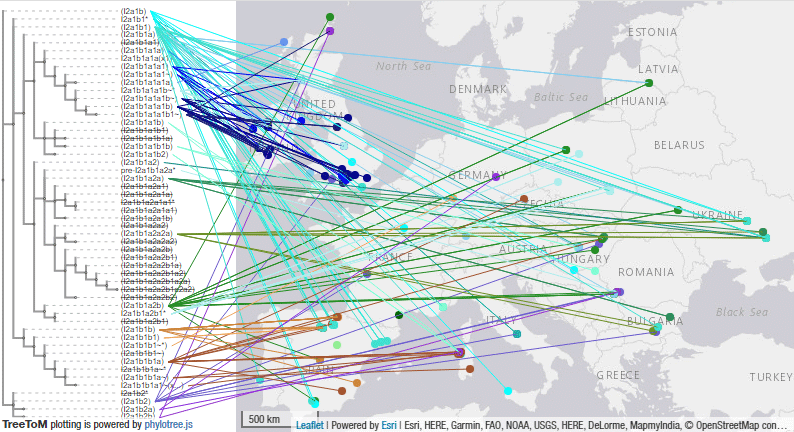
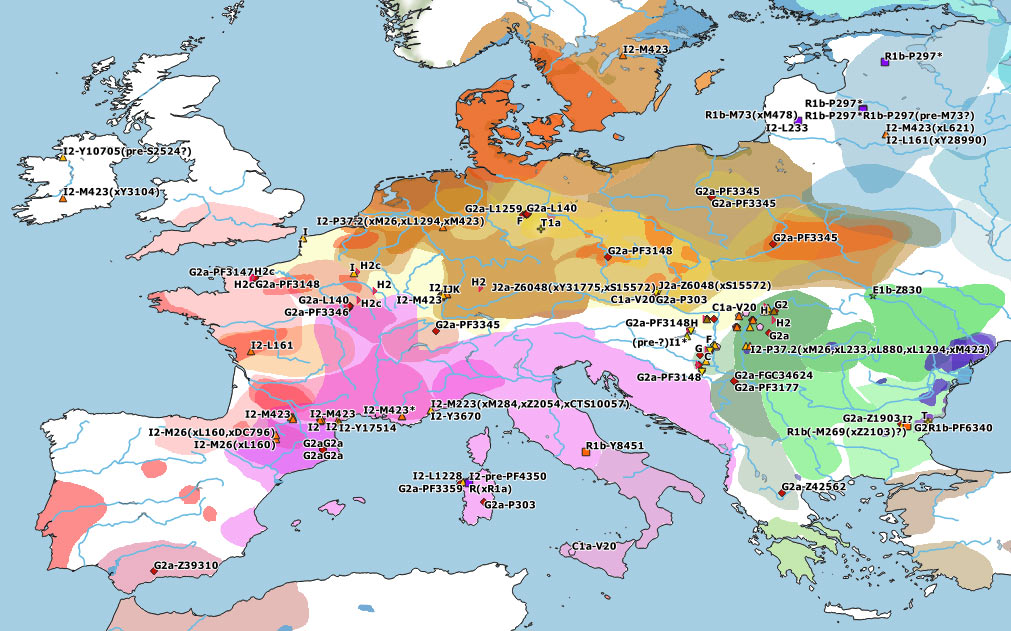
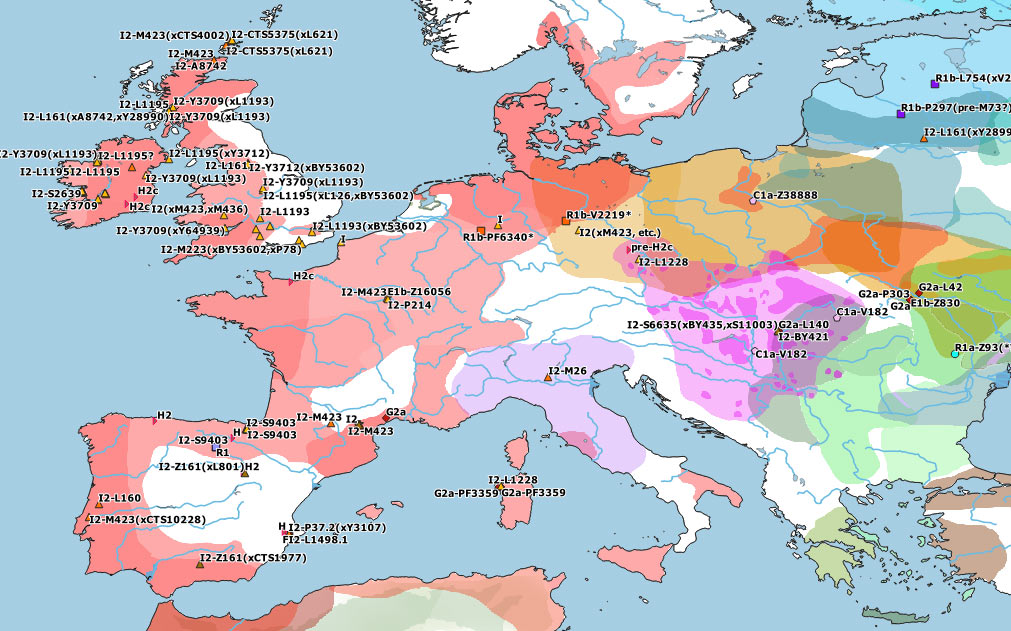
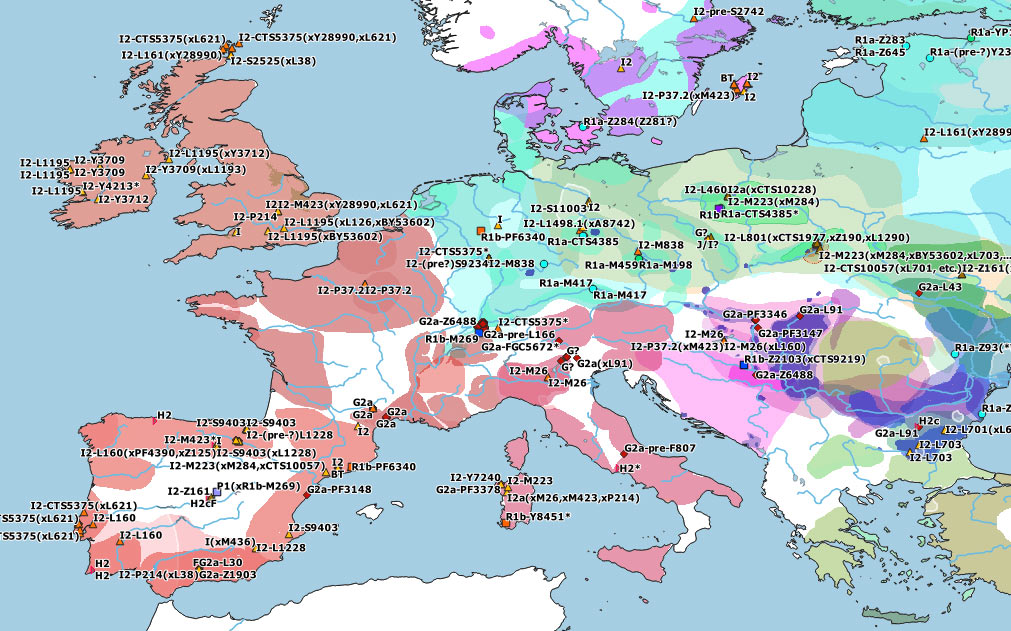
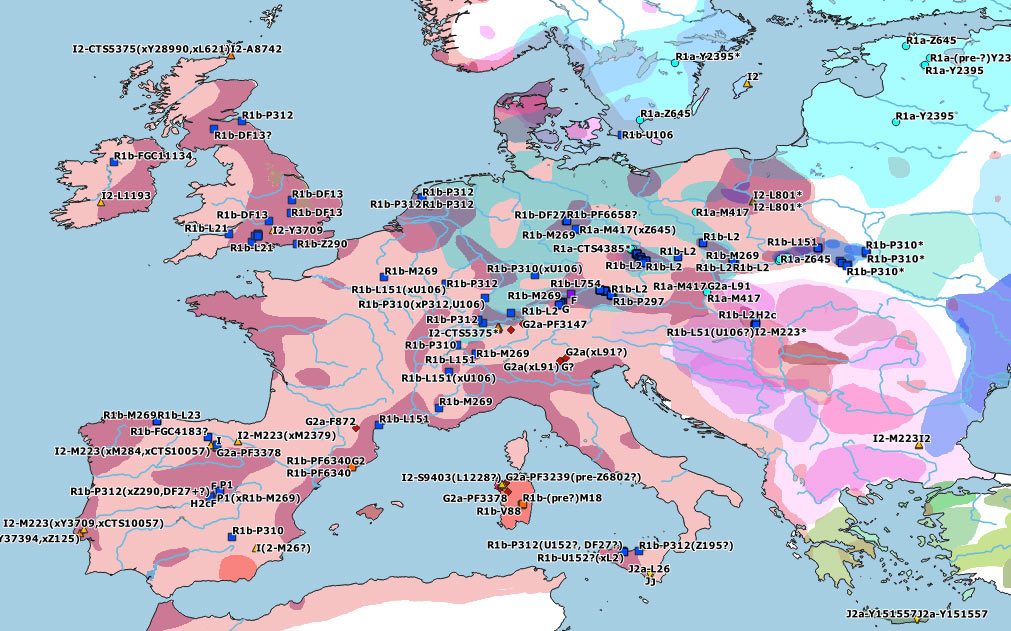
These are some TreeToM phylogeographic maps I have put together with the updated haplogroups reported in the paper, to replace the slightly outdated (and rather conservative) maps published in Cassidy et al. (2020), after shallowly reviewing SNP calls of those reported by them and previously reported ones (click on each one to view the dynamic version):
NOTE. For detailed information on each sample and haplogroup (and correspondence to ISOGG 2019-2020 nomenclature) you can check out the Ancient DNA Dataset, as well as the Arcgis Online Web App for their geographic distribution. Both are accessible at haplogroup.info.
I2a1a1-CTS595 subclades
I2a1a2-M423 subclades
I2a1b-M436 subclades
Comments
Feminist Old Europe?
The idea of egalitarian, peaceful, matriarchal, matrifocal (and matrilineal?) societies in Old Europe before the arrival of Steppe-related migrations is crumbling with each new report of ancient DNA, much like Gimbutas’ correlative simplistic idea of macho Kurgan peoples. Not only do “pre-complex” societies show a strong patriarchal structure and strict patrilineal descent, but, based on recently reported papers (see e.g. Beau et al. 2017 or Alt et al. 2020), they seem to be capable of unhinged violence like any other throughout history, which in hindsight should have been the most reasonable assumption about our stateless human past.
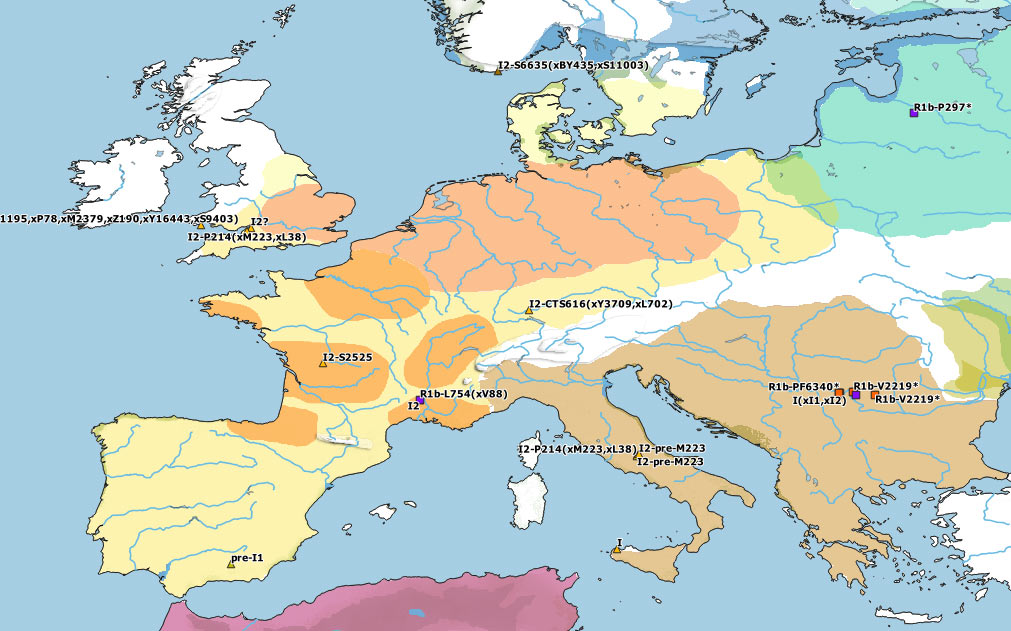
Neolithisation wave or waves?
The Neolithisation process in the Near East certainly shows increasing societal complexity: the incorporation of different hunter-gatherer (HG) haplogroups during demic diffusion events, evidenced by the increase and decrease in haplogroup variability depending on the specific sampled group and period – despite a generalized large continuity of ancestry – suggests that farmer populations were demographically stronger and that they had social arrangements allowing for female (and possibly male) exogamy, which were likely combined with subsequent changes in power structures.
This increasing complexity attested brings about an interesting theoretical question – but with important practical consequences – not dissimilar to the Ship of Theseus-like “Steppe ancestry” paradox: since ancestry remains mostly the same among Neolithic societies, but haplogroups evolve or radically change, does that mean that all early European farmer (EEF) groups spoke the same language? And, if not…when did they change it, and why?
That is, does the majority of North-West Anatolia Neolithic-like (NWAN) ancestry among expanding EEF cultures reveal a non-stop expansion of Anatolian farmers that somehow got hitchhiked through random Y-DNA bottlenecks, or did “local” groups of mainly HG paternal lineages occasionally dominate over demographically stronger farmer societies, imposing their language, and then expanded farther westward and northward?
The first Neolithic demic diffusion waves in Europe seem to have been dominated by G2a lineages, but they clearly also included haplogroups H2, E1b, J2, and T1a. Specific subclades of these haplogroups partook in the leveling of ancestry that happened in the Near East earlier than ca. 6500 BC, so it remains unclear how many different Near Eastern groups might have taken part in the different Neolithic waves of expansion into Europe and throughout the Mediterranean – the simplistic reference to NWAN ancestry notwithstanding.
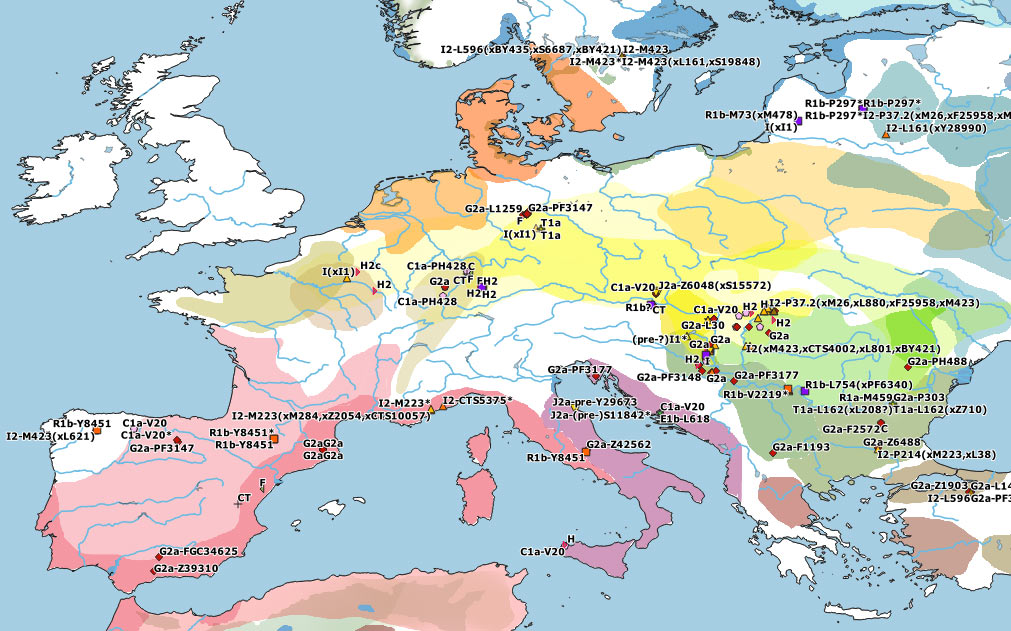
HG haplogroups vs. NWAN ancestry
Even more interesting than how many waves originated in Anatolia and/or nearby regions, and whether or not one could speak about a single “Anatolia Neolithic language”, is the incorporation of haplogroups during their stepped spread throughout Europe. For example, the southern Neolithic wave reached the Central Mediterranean with an incorporation of hg. R1b-V2219 – with a likely expansion from SE Europe – and its frequency seems to have peaked under R1b-V88 bottlenecks among farmers of Central Italy, Sardinia and Northern Iberia during the Early to Middle Neolithic.
Furthermore, in the Western Mediterranean and in Western Europe, HG-related I2a subclades became dominant, although it remains unclear where exactly each of these different subclades might have become incorporated to expanding EEF-like populations. And, even though isolated I2 samples show occasionally almost full HG-related ancestry, it seems to be a well-established general pattern that HG ancestry got diluted among the likely demographically denser farmer populations that swept over west- and northward throughout Europe, whether HG-related paternal lineages dominated over or were absorbed by NWAN-related ones.
Haplogroups alright, but what about Neandertals?
With each succeeding Neolithic diffusion wave, during the centuries (or millennia) that farming subsistence economy kept spreading, more haplogroups proper of European hunter-gatherers* became incorporated and spread further.
*some I2 subclades were also present in Anatolia, probably associated with Palaeolithic and Mesolithic expansions, so not all I2 subclades attested have necessarily a direct origin in Europe.
These simplistically called resurgence events mean that communities with “native” WHG- or EHG-related male lineages either:
a) became part of expanding farming communities through some socially acceptable mechanism, as it seems to be the case, for example, among sampled late disintegrating Corded Ware groups of the mid-3rd millennium BC; or
b) (much more likely) demographically weak hunter-gatherer communities eventually dominated over some expanding Neolithic groups and their ancestry was diluted among the prevalent EEF-related ancestry through admixture – much like Proto-Indo-Europeans seem to have been originally EHG-like, but admixed with peoples of so-called “Steppe ancestry”, or much like Proto-Anatolians seem to have been originally Steppe-like and rich in hg. R1b, but Common Anatolians likely resembled surrounding Near Eastern populations.
On the other hand, driving the haplogroup-based definition of linguistic groups ad absurdum leads us again to the excesses of the 1990s: if a “majority haplogroup” defines a culture despite ancestry, then what about Neandertals? Some of them – possibly a majority of them just before their extinction – show male and female lineages proper of Anatomically Modern Humans (see Petr et al. 2020, contra Mendez et al. 2016). So when does a Neandertal community actually become an Anatomically Modern Human one? Is it ancestry, haplogroup, behaviour (culture)…or a combination of them? And if it is a combination, which is more important, and where is the threshold?
Language controversy
Before I digress further, the target question is – what defines language evolution or language change? Haplogroup, ancestry, culture? A combination of them? And if so, which one? I know that a lot of people can’t work with hg. A000 among Neandertals in their minds (some can’t even put R1b-L51 and R1b-Z2103 together), so let me rephrase it in a culturally appropriate setting: if Neandertals had survived secluded in some mountainous region up until the Neolithic and showed haplogroup R1a…were they Indo-European speakers? If they were of hg. N1c…were they Uralic speakers? Would it depend on how much “Steppe ancestry” or “Siberian ancestry” they had on top of Neandertal ancestry? Would it depend on whether they were sampled in the Eastern Carpathians or in the Urals?
For most people – surprisingly, even for many interested in the prehistory of the Basque language but still obsessing over an impossible origin of hg. R1b-P312 – papers like this one aren’t relevant at all: for some reason, whereas these Neolithic populations are considered dead long ago, others (who became also extinct) appear more attractive from the perspective of ethnic- and nation-building myths. Largely uncontroversial periods like the Megalithic culture might offer thus an interesting opportunity that could be used to explore alternatives and to interpret subsequent genomic, cultural and linguistic processes that happened during the much more contentious Steppe-related migrations, whose interpretation is thus prone to strong biases.
Afroasiatic vs. Vasconic
Let’s assume that Anatolia Neolithic spread Afroasiatic during the 6th millennium BC, which is currently – after the report of Prendergast et al. (2019) – the most parsimonious explanation of the population expansions that must have taken place in Northern Africa for the language to spread just before the end of the Green Sahara period. Vasconic (potentially Basque-Iberian) must have been spoken in Western Europe at the arrival of North-West Indo-European-speaking Bell Beakers. So the question is: when and how did Vasconic (or any other pre-Neolithic language group) survive the massive NWAN-related waves?
For example, assuming that hg. R1b-V88 spread through the Central Mediterranean into Northern Africa linked to Afroasiatic languages during the Early Neolithic expansions, and that Paleosardo is closely related to Iberian – hence possibly part of a later HG “resurgence” event – it seems that both hg. I2a1b1b-Y6098 and I2a1a1a-M26 are the only potential (I mean evident, excluding more complex) proxies for a HG-related language survival and its expansion after the first Neolithic waves and until the arrival of Yamnaya-derived East Bell Beakers. But when and how did that survival and expansion happen? Was it rather late, with megaliths? with Proto-Beakers (some of whom also show hg. R1b-V88, like ancient Sardinians)? even later through subtler (cultural) diffusion events? And would a common Iberian-Sardinian ancestry matter that much (see Marcus et al. 2019)? More or less than common haplogroups?
Also, how relevant for a linguistic argument would it be that I2-rich societies showed more or less HG-related ancestry? Because, as far as we know, that kind of generally described ancestral component keeps popping up until well into the Late Neolithic in Western Europe, so it’s difficult if not impossible to find an uninterrupted trace of some specific Mesolithic HG-related ancestry (e.g. a specific Villabruna ± ElMirón combination) surviving unscathed through post-Neolithic times…And what about groups showing the occasional NWAN-related Y-DNA bottleneck – like H2 in this paper, or G2a, E1b in other parts of Western or Central Europe? Do any of them challenge the nature of the language spoken by I2-rich groups?
The “precise homeland” fallacy
Some subclades are occasionally so well documented during succeeding explosive and/or massive expansions that we can trace their origin quite accurately, despite changes in ancestry. R1b-L23 and R1a-M417 are two of those cases, for obvious reasons. But even in cases where haplogroup and ancestry show a tight connection, the doubt remains: where did this or that group actually stem from? I mean, literally, where exactly were their core settlements, where was their homeland? Which group among the many described (and the many unknown) is the direct ancestral population of those sampled?
Such a demand for accuracy is impossible to answer, and it’s the kind of question usually posed for spurious reasons, to challenge what is becoming well established. As an example, we know that the formation of Pre-Yamnaya ancestry coincided with the expansion of R1b-rich Khvalynsk chieftains, which were probably still mostly EHG-like in the Middle Volga area before the leveling of their ancestry through exogamy during their expansion into the Lower Volga and Northern Caucasus steppes. To keep looking for “Steppe ancestry” similar to Yamnaya to locate the Proto-Indo-European homeland against the already known cultural expansion, linguistic estimates, and findings in population genomics reveals a fallacious selective perception as to what population genomics is able to prove, and what it has in fact already proven.
NOTE. Feel free to comment below if you know of a clear geographical+genetic connection from one precise group to another separated in time by a span of 1,000+ years, anywhere in the world in prehistoric times, which could be used to establish a direct cultural-historical connection. The only ones that come to mind are the Yamnaya – Bell Beaker connection, and the Bronze to Iron Age continuity of the Eastern Baltic, and even those are usually doubted for the most varied and colorful reasons…
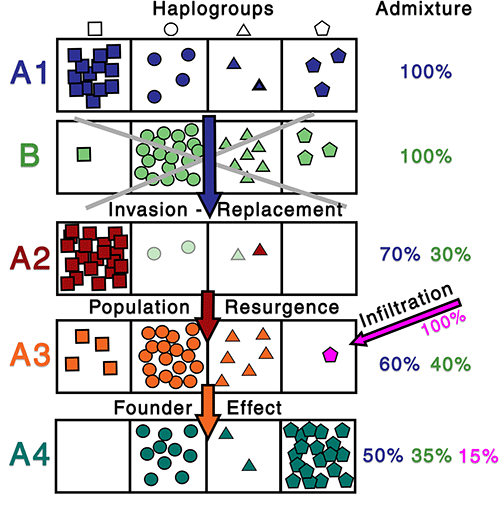
A theoretical framework?
For example, in the case of Early Neolithic farmers – meaning early SE European, LBK, Impresso-Cardial groups with a majority of Near Eastern haplogroups:
Can we say that all Anatolia-related groups expanding into Europe actually originated in one roughly limited region close to ca. 7000-6500 BC, just because they show an apparently common “NWAN-related ancestry”?
If we assume that NWAN was a single ancestral component dated to ca. 7000 BC and found in a single region, and that it was completely different from other neighbouring groups… does that mean that all expanding groups (despite their different haplogroups, genetic and cultural interactions) had the same, a similar, or even related languages?
Could they represent all (most of them unrelated) non-Indo-European languages of Europe, attested millennia later? And why, exactly? To justify a native Vasconic Europe, a native Etruscan-speaking Italy?
In case you haven’t noticed, population genomics has a clear limit as to what it can explain alone, and every single cultural change coupled with the intrusion of haplogroups and ancestry changes seems to offer, instead of closure, an opportunity, a void canvas where one can disregard all other data and let their imagination go wild. Do you find a 99% A vs. 1% B ancestry? Maybe the 1% B is your connection, depending on what you were set out to prove. You find then an 80% B vs. 20 % C ancestry? The 80% B might be now what matters, even though the 1% was very important before…
Better still, do you find the expected B-related haplogroup(s) but no B ancestry? That can be explained by great dilution events and the haplogroup is key to show the link. Alternatively, in case you want to dismiss an uncomfortable link, you can speak of infiltration and cultural diffusion, so a majority of B-related lineages (even coupled with B-related ancestry) doesn’t matter anymore…
More relevant questions to understand cultural evolution are:
- Whether ancestry B was:
- exclusive of a well-defined population, related chronologically, geographically, and culturally to the hypothetically ancestral one; or
- part of a continuum of similar ancestries that may well have been spread over a vast area, among completely different cultural groups, for thousands of years.
- Whether the SNPs defining B-related subclades – as suggested by their estimated formation date and TMRCA – are coincident or compatible with the date of the cultural spread with which they might have been associated, hence supporting the hypothesized demic diffusion event(s).
God-kings and other citebait
The point of this rant – which occurred to me while reviewing countless SNP calls for I2 subclades in the Dataset, unable to find a clear connection of continuous demic diffusion events among Megalithic societies – is this: do researchers ever stop to think about setting some ground rules before reaching conclusions? You know, like having explicit cultural models of how reconstructed languages might relate to the spread of prehistoric cultures, and how these are coupled with (or decoupled from) genomic descent, and what ancestry and haplogroups actually mean? Do they then try to fit that with what they find? Or is population genomics becoming a game of ad hoc conclusions, were the antecedent is the result and data is useful only to liven up the discussion?
The far-fetched reference in Cassidy et al. (2020) to god-kings (in the abstract) and Celtic folklore (in the content) regarding a Megalithic culture that was replaced many times over in Ireland, and a cultural taboo that is found in mythological accounts worldwide, suggests that Nature editors and reviewers were keen on keeping those references. I can’t subscribe the criticism against authors, because it would have been really easy to strike those references out at any point during the review process, so everybody involved is to blame. The single mention of “god-king” in the abstract and the lack of discussion about it in the paper seems to me enough proof that editors assumed most people would not read past it.
Some are now mocking the “Celtic megalith-builder incestuous pharaohs”. Sure, why not, let’s have fun defining what is and isn’t science. But the truth is that most papers in population genomics as they are linked to culture, and especially to prehistoric languages and language evolution, engage in similarly risible eye-catching headlines…And I find this self-evident PR stunt of the Celtic Targaryen in an overall well-balanced and well-written paper far less damaging to science than the constant fiascos of apparently subtler over-stated conclusions that continue to plague the field.
You can’t have your “strict science” cake and also eat it.
The featured image of this post is Detlef Gronenborn’s Map: Expansion of farming in western Eurasia, 9600 – 4000 cal BC (update vers. 2019.3).
See also
- Survival of hunter-gatherer ancestry in West-Central European Neolithic
- Demographically complex Near East hints at Anatolian and Indo-Aryan arrival
- Visualizing phylogenetic trees of ancient DNA in a map
- Spread of Indo-European and Uralic speakers in ADMIXTURE
- Volga Basin R1b-rich Proto-Indo-Europeans of (Pre-)Yamnaya ancestry
- “Steppe ancestry” step by step (2019): Mesolithic to Early Bronze Age Eurasia
- Bell Beakers and Mycenaeans from Yamnaya; Corded Ware from the forest steppe
- Yamnaya ancestry: mapping the Proto-Indo-European expansions
- Corded Ware ancestry in North Eurasia and the Uralic expansion