The definitive publication of a BioRxiv preprint article, in Nature: Parallel palaeogenomic transects reveal complex genetic history of early European farmers, by Lipson et al. (2017).
The dataset with all new samples is available at the Reich Lab’s website. You can try my drafts on how to do your own PCA and ADMIXTURE analysis with some of their new datasets.
Abstract:
Ancient DNA studies have established that Neolithic European populations were descended from Anatolian migrants who received a limited amount of admixture from resident hunter-gatherers. Many open questions remain, however, about the spatial and temporal dynamics of population interactions and admixture during the Neolithic period. Here we investigate the population dynamics of Neolithization across Europe using a high-resolution genome-wide ancient DNA dataset with a total of 180 samples, of which 130 are newly reported here, from the Neolithic and Chalcolithic periods of Hungary (6000–2900 BC, n = 100), Germany (5500–3000 BC, n = 42) and Spain (5500–2200 BC, n = 38). We find that genetic diversity was shaped predominantly by local processes, with varied sources and proportions of hunter-gatherer ancestry among the three regions and through time. Admixture between groups with different ancestry profiles was pervasive and resulted in observable population transformation across almost all cultural transitions. Our results shed new light on the ways in which gene flow reshaped European populations throughout the Neolithic period and demonstrate the potential of time-series-based sampling and modelling approaches to elucidate multiple dimensions of historical population interactions.
There were some interesting finds on a regional level, with some late survival of hunter-gatherer ancestry (and Y-DNA haplogroups) in certain specific sites, but nothing especially surprising. This survival of HG ancestry and lineages in Iberia and other regions may be used to revive (yet again) the controversy over the origin of non-Indo-European languages of Europe attested in historical times, such as the only (non-Uralic) one surviving to this day, the Basque language.
This study kept confirming the absence of Y-DNA R1b-M269 subclades in Central Europe before the arrival of Yamna migrants, though, which offers strong reasons to reject the Indo-European from the west hypothesis.
Here are first the PCA of samples included in this paper, and then the PCA of ancient Eurasians (Mathieson et al. 2017) and modern populations (Lazaridis et al. 2014) for comparison of similar clusters:
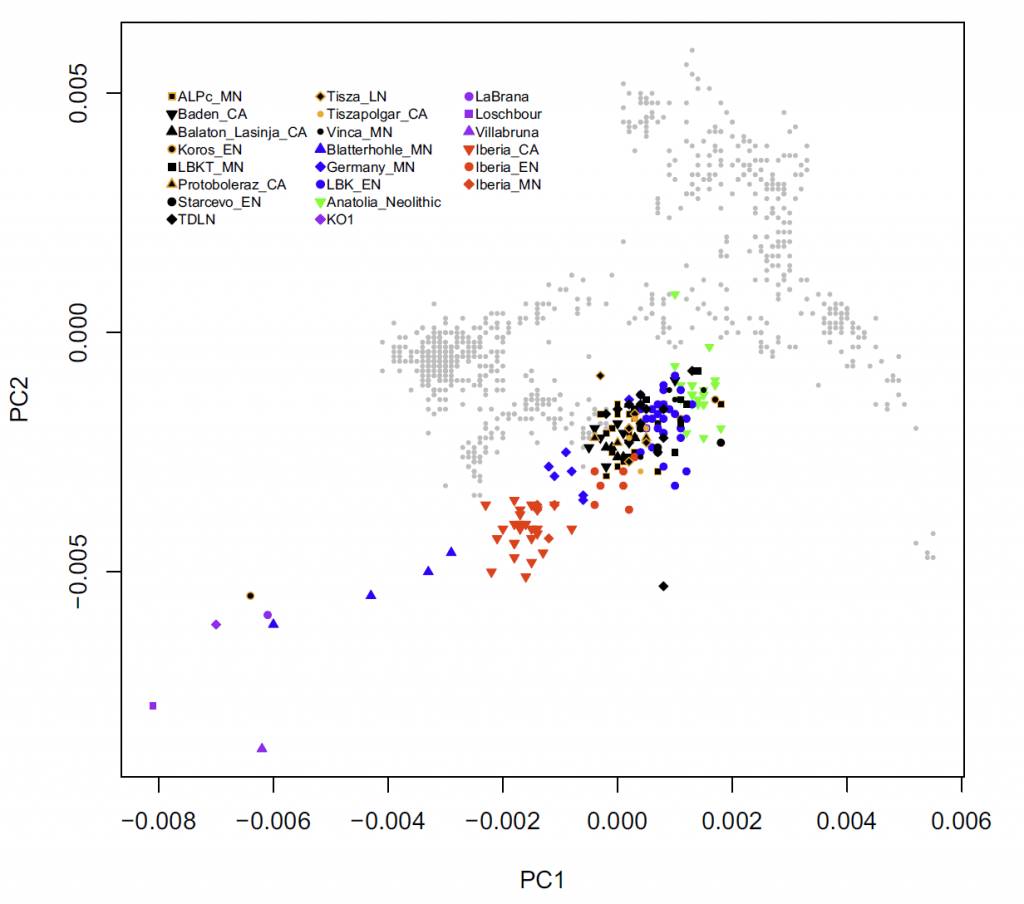
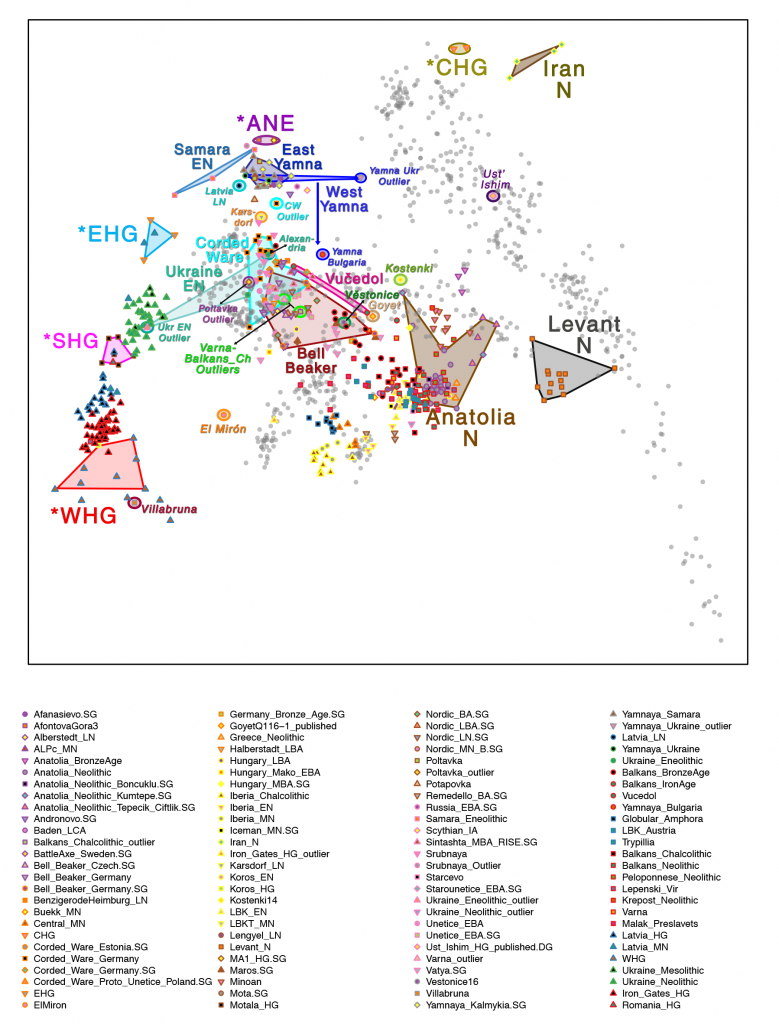
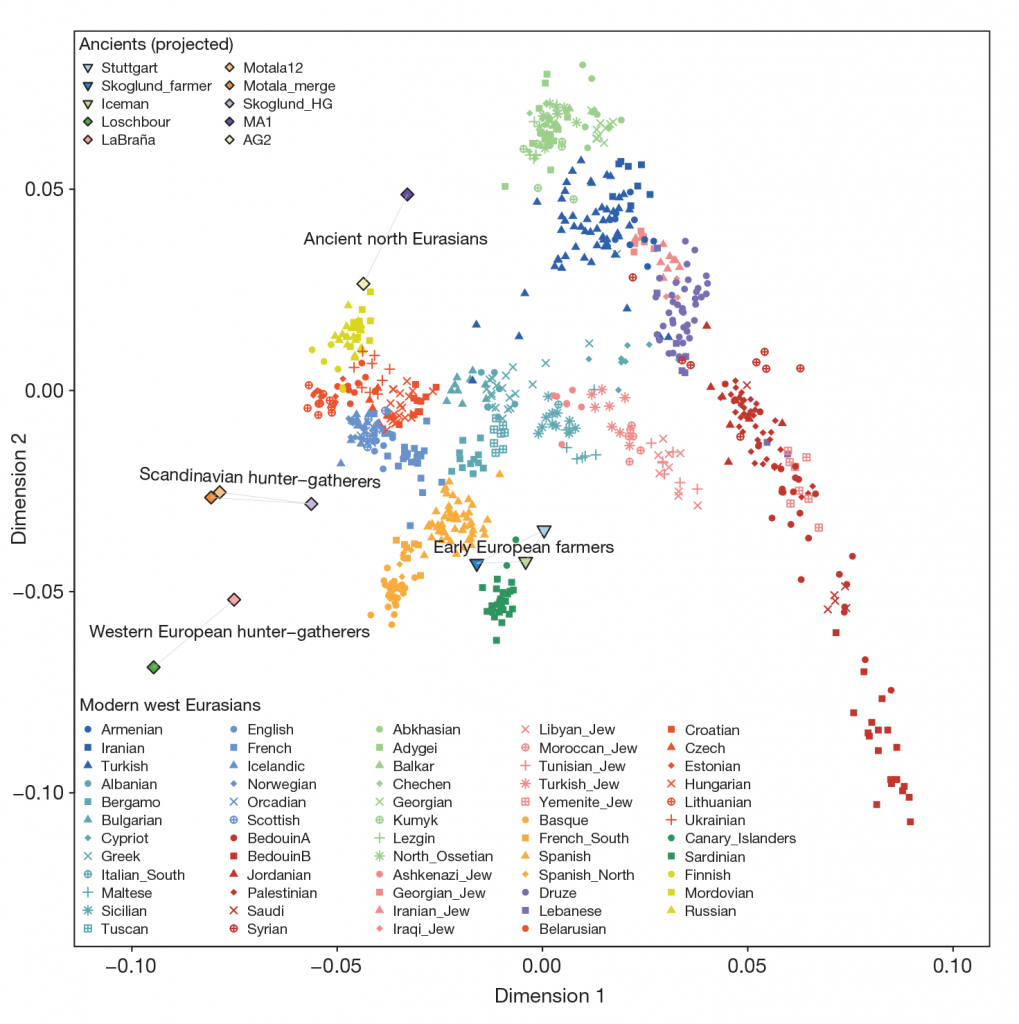
Related:
- Human ancestry: how to work your own PCA, ADMIXTURE analyses for human evolutionary and genealogical studies
- Iberian Peninsula: Discontinuity in mtDNA between hunter-gatherers and farmers, not so much during the Chalcolithic and EBA
- Analysis of R1b-DF27 haplogroups in modern populations adds new information that contrasts with ‘steppe admixture’ results
- Genetic origins of Minoans and Mycenaeans and their continuity into modern Greeks
- Neolithic and Bronze Age Basque-speaking Iberians resisted invaders from the steppe
- How to do modern phylogeography: Relationships between clans and genetic kin explain cultural similarities over vast distances