New paper (behind paywall) Reconciling the father tongue and mother tongue hypotheses in Indo-European populations, by Zhang et al. National Science Review (2018) nwy083.
Interesting excerpts:
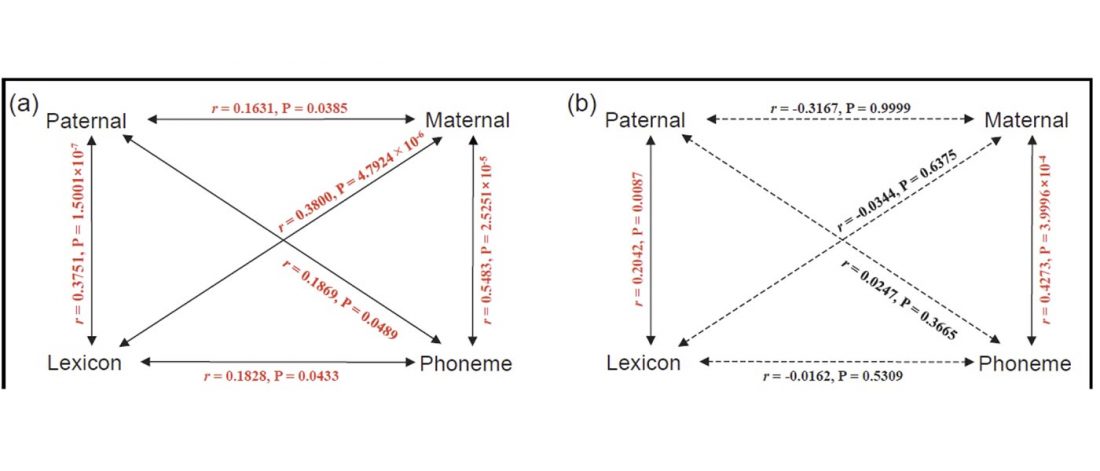
Here, we reassessed the correlation between genetic and linguistic characteristics in 34 modern IE populations (Fig. 1a), for which all four types of datasets (lexicon, phonemes, Y-chromosomal composition, and mitochondrial DNA (mtDNA) composition) are available. We assembled compositions of the Y-chromosomal and mtDNA haplogroups or paragroups from the corresponding IE populations, which reflect paternal and maternal lines, respectively (…)
… Read the rest “The father tongue and mother tongue hypotheses in Indo-European populations”Neighbour-Nets were constructed to delineate the differences between 34 IE population groups clustering at