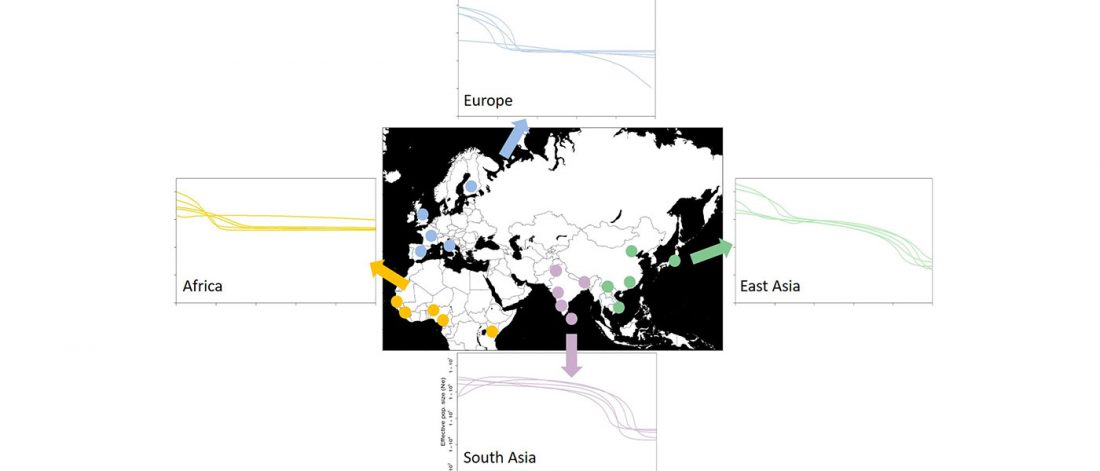
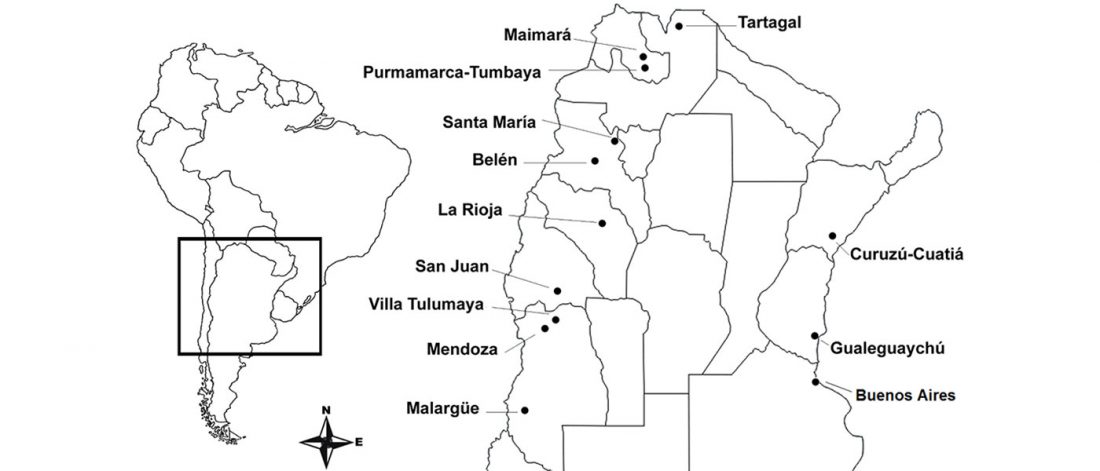
Open access Global demographic history of human populations inferred from whole mitochondrial genomes, by Miller, Manica, and Amos, Royal Society Open Science (2018).
Relevant excerpts (emphasis mine):
Material
… Read the rest “Global demographic history inferred from mitogenomes”The Phase 3 sequence data from 20 populations, comprising five populations for each of the four main geographical regions of Europe, East Asia, South Asia and Africa, were downloaded from the 1000 Genomes Project website (www.1000genomes.org/data, [8]), including whole mitochondrial genome data for 1999 individuals. We decided not to analyse populations from the Americas due to the region’s complex history of admixture [13,14].
The European populations were as follows: Finnish sampled