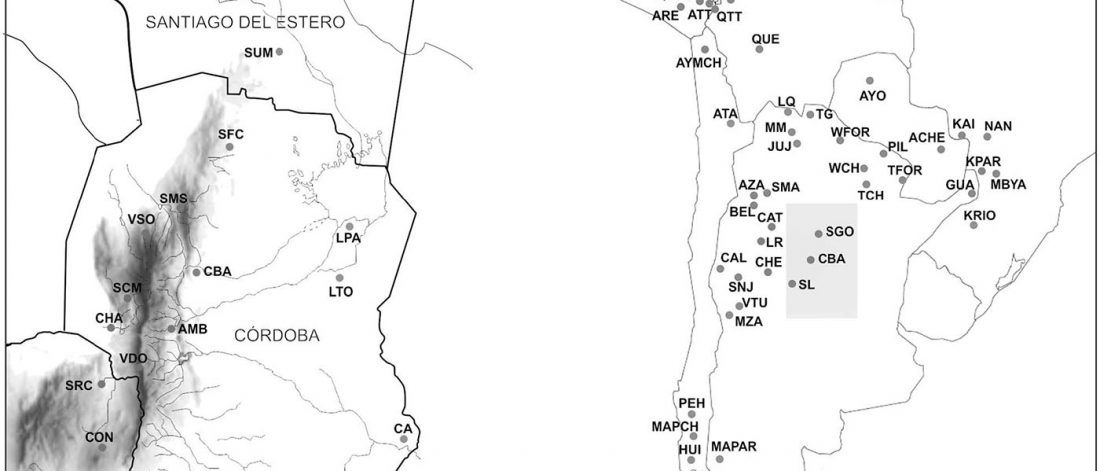
Paternal lineages mainly from migrants, maternal lineages mainly from local populations in Argentina
New paper (behind paywall) Genetic variation in populations from central Argentina based on mitochondrial and Y chromosome DNA evidence, by García, Pauro, Bailliet, Bravi & Demarchi, J. Hum. Genet (2018) 63: 493–507.
Abstract (emphasis mine):
… Read the rest “Paternal lineages mainly from migrants, maternal lineages mainly from local populations in Argentina”We present new data and analysis on the genetic variation of contemporary inhabitants of central Argentina, including a total of 812 unrelated individuals from 20 populations. Our goal was to bring new elements for understanding micro-evolutionary and historical processes that generated the genetic diversity of the region, using molecular markers of uniparental inheritance (mitochondrial DNA and Y chromosome). Almost 76% of the individuals show