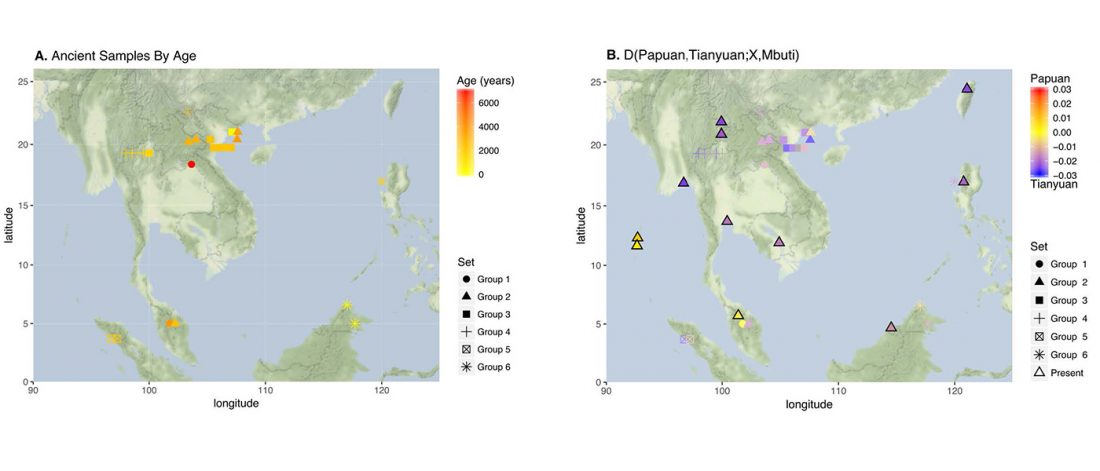
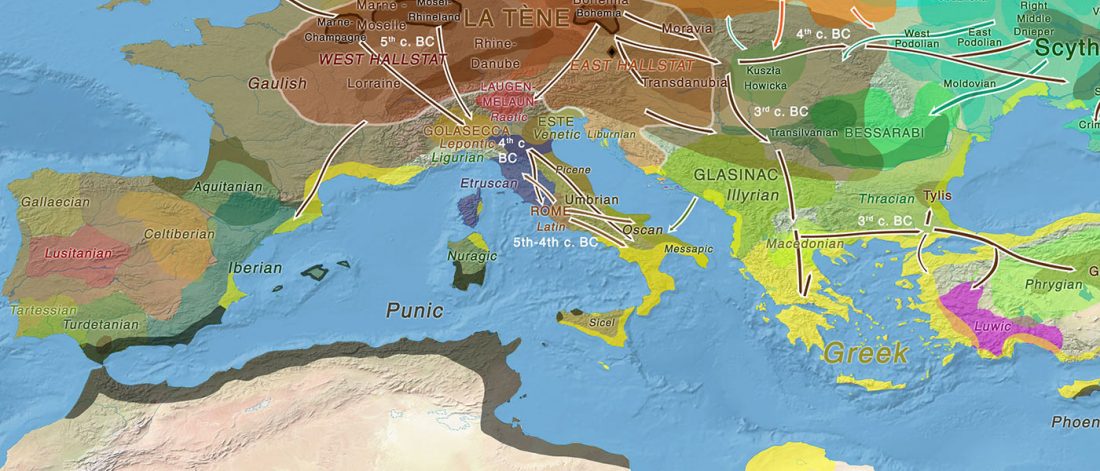
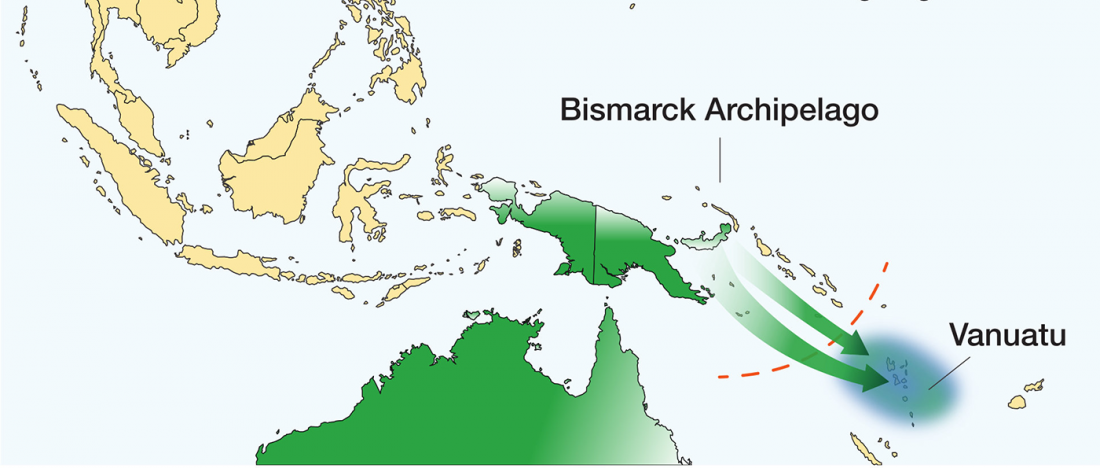
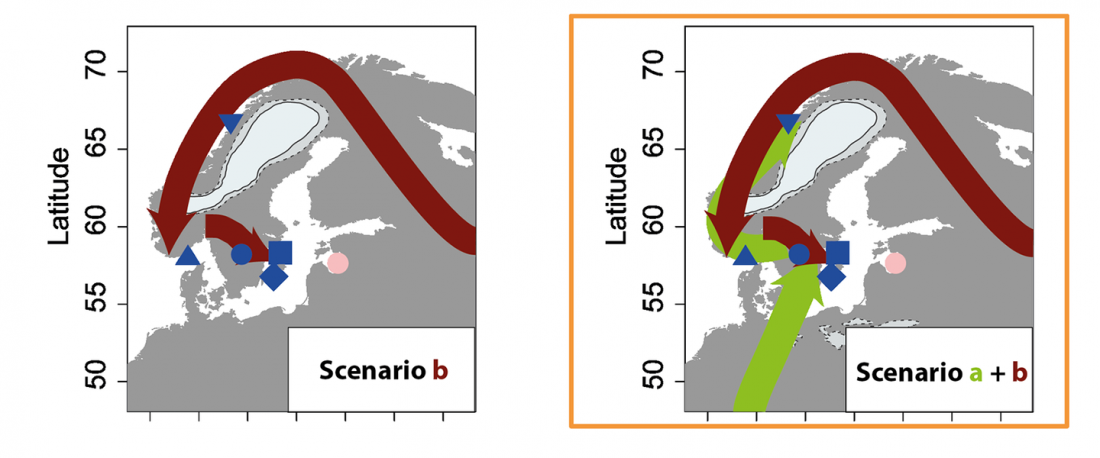
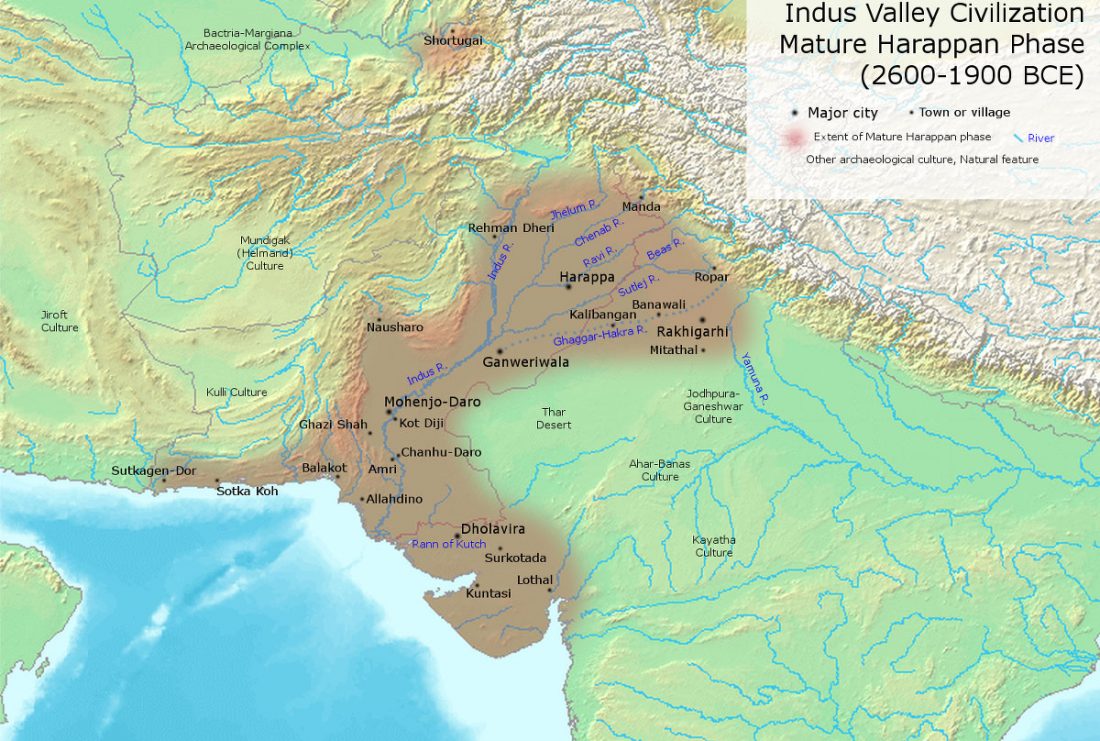
Genomics reveals four prehistoric migration waves into South-East Asia
Open access preprint article at bioRxiv Ancient Genomics Reveals Four Prehistoric Migration Waves into Southeast Asia, by McColl, Racimo, Vinner, et al. (2018).
Abstract (emphasis mine):
… Read the rest “Genomics reveals four prehistoric migration waves into South-East Asia”Two distinct population models have been put forward to explain present-day human diversity in Southeast Asia. The first model proposes long-term continuity (Regional Continuity model) while the other suggests two waves of dispersal (Two Layer model). Here, we use whole-genome capture in combination with shotgun sequencing to generate 25 ancient human genome sequences from mainland and island Southeast Asia, and directly test the two competing hypotheses. We find that early genomes from Hoabinhian