Open access Emergence and Spread of Basal Lineages of Yersinia pestis during the Neolithic Decline, by Rascovan et al. Cell (2018)
Abstract (emphasis mine):
Between 5,000 and 6,000 years ago, many Neolithic societies declined throughout western Eurasia due to a combination of factors that are still largely debated. Here, we report the discovery and genome reconstruction of Yersinia pestis, the etiological agent of plague, in Neolithic farmers in Sweden, pre-dating and basal to all modern and ancient known strains of this pathogen. We investigated the history of this strain by combining phylogenetic and molecular clock analyses of the bacterial genome, detailed archaeological information, and genomic analyses from infected individuals and hundreds of ancient human samples across Eurasia. These analyses revealed that multiple and independent lineages of Y. pestis branched and expanded across Eurasia during the Neolithic decline, spreading most likely through early trade networks rather than massive human migrations. Our results are consistent with the existence of a prehistoric plague pandemic that likely contributed to the decay of Neolithic populations in Europe.
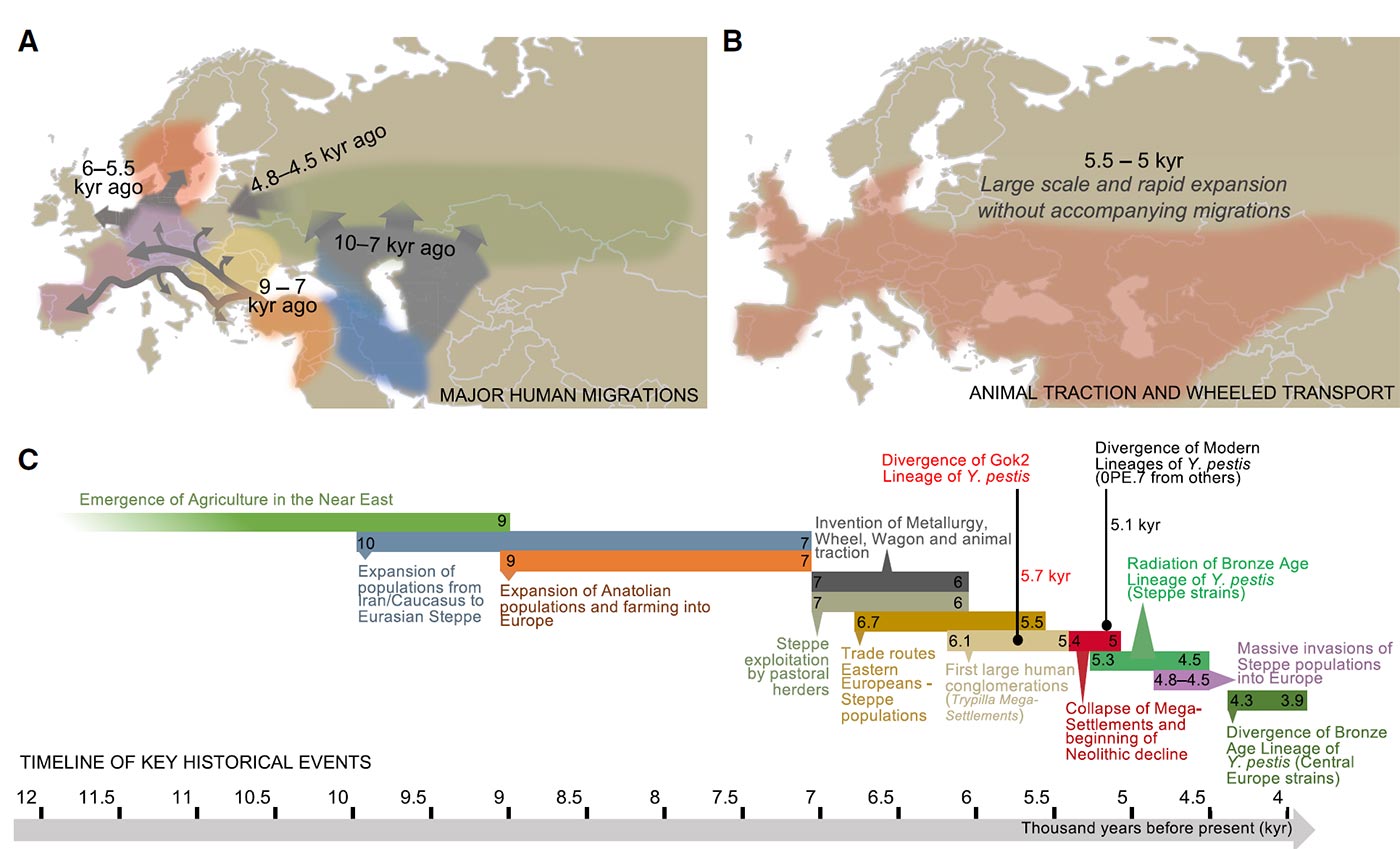
(B) Geographic distribution of the use of animal traction and wheeled transport across Neolithic and Bronze Age populations in Eurasia, which broadly expanded during the period of 5,500 and 5,000 BP. The expansion of these technological innovations overlaps the predicted period for the expansion of the basal Y. pestis strains.
(C) Timeline indicating the proposed key historical events that contributed to the emergence and spread of plague during the Neolithic.
We have evolved in the interpretation of the plague from 1) a Corded Ware-driven disease, to 2) a steppe disease that was spread by Yamna and Corded Ware, and now 3) a (potentially) Trypillia-driven disease that spread to the west earlier than Yamna and Corded Ware, but probably also later east and west with both.
At least it still seems that the plague and its demographic consequences were a good reason for the expansion of Indo-Europeans and Uralians into Europe, as we thought…
Featured image, from the paper: “The predicted model of early dispersion of Y. pestis during Neolithic and Bronze Age was built by integrating phylogenetic information of Y. pestis strains from this period (Figure 1E), their divergence times (Figure 3), the geographic locations, carbon dating and genotypes of the individuals, and the archaeological record. The model suggests that early Y. pestis strains likely emerged and spread from mega-settlements in Eastern Europe (built by the Trypillia Culture) into Europe and the Eurasian steppe, most likely through human interaction networks. This was facilitated by wheeled and animal-powered transports, which are schematized in the map with red lines with arrows pointing in both senses. Our model builds upon a previous model (Andrades Valtuena et al., 2017) that proposed the spread of plague to be associated with large-scale human migrations (blue line).”
Related
- Cystic fibrosis probably spread with expanding Bell Beakers
- Oldest bubonic plague genome decoded in Srubna ca. 3800 YBP
- Phylogeny of leprosy, relevant for prehistoric Eurasian contacts
- Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans
- Stone Age plague accompanying migrants from the steppe, probably Yamna, Balkan EBA, and Bell Beaker, not Corded Ware
- The Caucasus a genetic and cultural barrier; Yamna dominated by R1b-M269; Yamna settlers in Hungary cluster with Yamna
- Steppe and Caucasus Eneolithic: the new keystones of the EHG-CHG-ANE ancestry in steppe groups
- East Bell Beakers, an in situ admixture of Yamna settlers and GAC-like groups in Hungary