Open access paper Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture, by Sharon L. Browning, Brian L. Browning, Zhou, Tucci, & Akey, Cell (2018).
Summary:
Anatomically modern humans interbred with Neanderthals and with a related archaic population known as Denisovans. Genomes of several Neanderthals and one Denisovan have been sequenced, and these reference genomes have been used to detect introgressed genetic material in present-day human genomes. Segments of introgression also can be detected without use of reference genomes, and doing so can be advantageous for finding introgressed segments that are less closely related to the sequenced archaic genomes. We apply a new reference-free method for detecting archaic introgression to 5,639 whole-genome sequences from Eurasia and Oceania. We find Denisovan ancestry in populations from East and South Asia and Papuans. Denisovan ancestry comprises two components with differing similarity to the sequenced Altai Denisovan individual. This indicates that at least two distinct instances of Denisovan admixture into modern humans occurred, involving Denisovan populations that had different levels of relatedness to the sequenced Altai Denisovan.
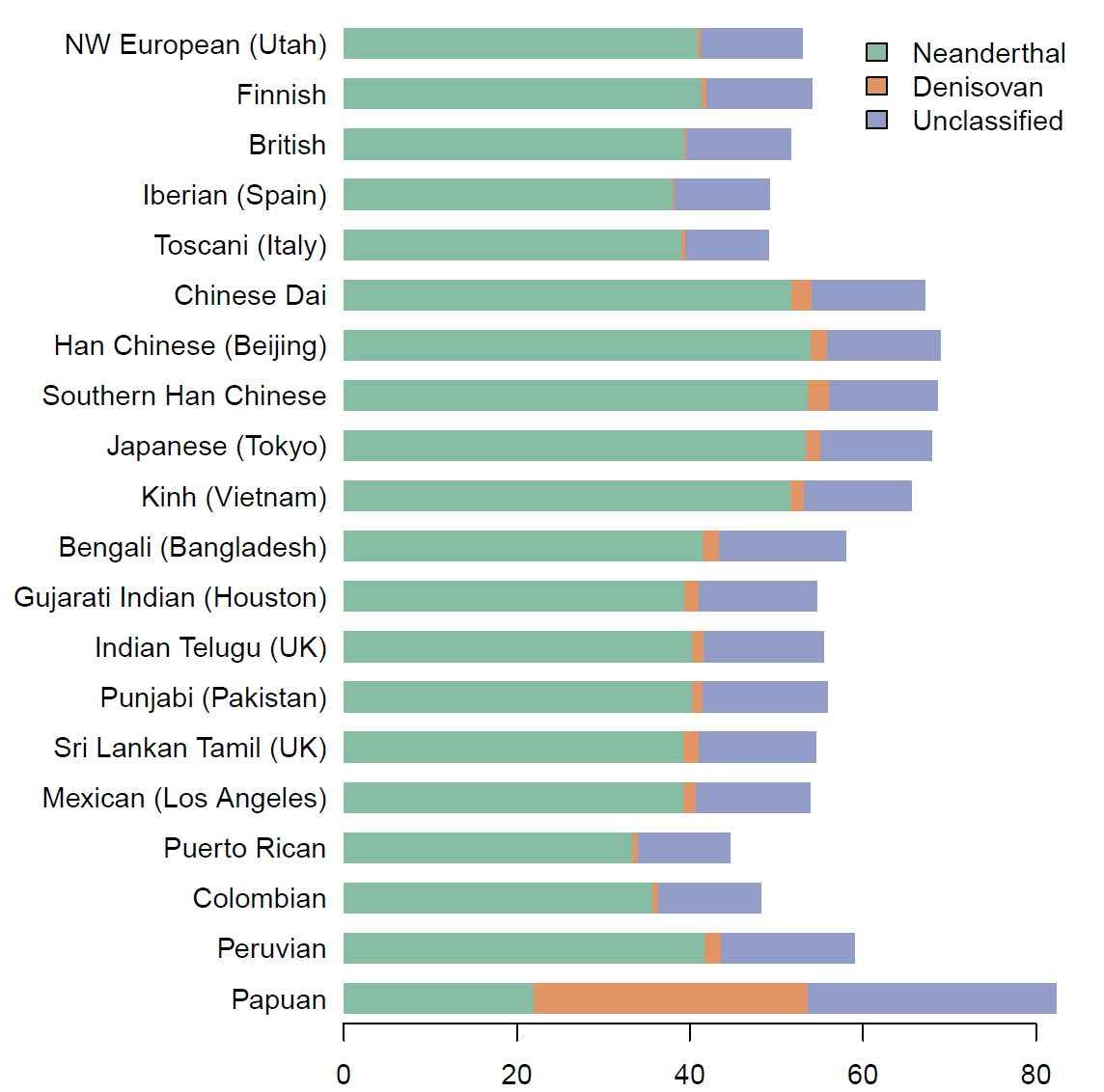
The discussion on the potential implication of the paper:
There is an undiscussed but potentially explosive implication of this paper: the 2nd Denisovan gene flow signal in East Asia seems to be absent from Native American ancestry–could Denisovans have survived after the isolation of these lineages <30 kya?? https://t.co/q348Z1iHXB pic.twitter.com/stWVJVObMQ
— Pontus Skoglund (@pontus_skoglund) March 15, 2018
Featured image, from the article: Contour Density Plots of Match Proportion of Introgressed Segments to the Altai Neanderthal and Altai Denisovan Genomes.
Related:
- Review article about Ancient Genomics, by Pontus Skoglund and Iain Mathieson
- Review article on the origin of modern humans: the multiple-dispersal model and Late Pleistocene Asia
- Ancient genomes document multiple waves of migration in south-east Asian prehistory
- Genomics reveals four prehistoric migration waves into South-East Asia
- Population turnover in Remote Oceania shortly after initial settlement
- Genomic history of South-East Asia: eastern Polynesians, Peninsular Malaysia and North Borneo
- Islands across the Indonesian archipelago show complex patterns of admixture
- Two more studies on the genetic history of East Asia: Han Chinese and Thailand
- Reconstructing the demographic history of the Himalayan and adjoining populations
- Ancient Di-Qiang people show early links with Han Chinese