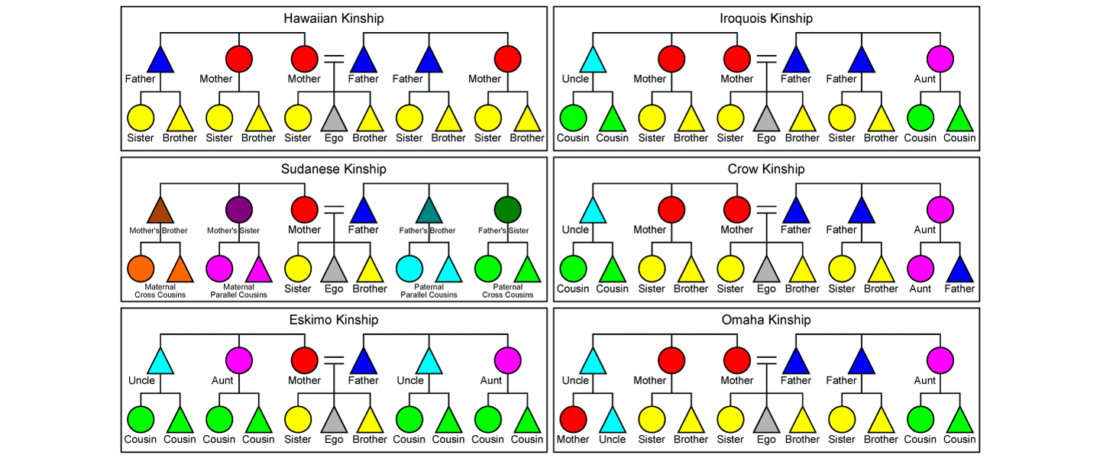
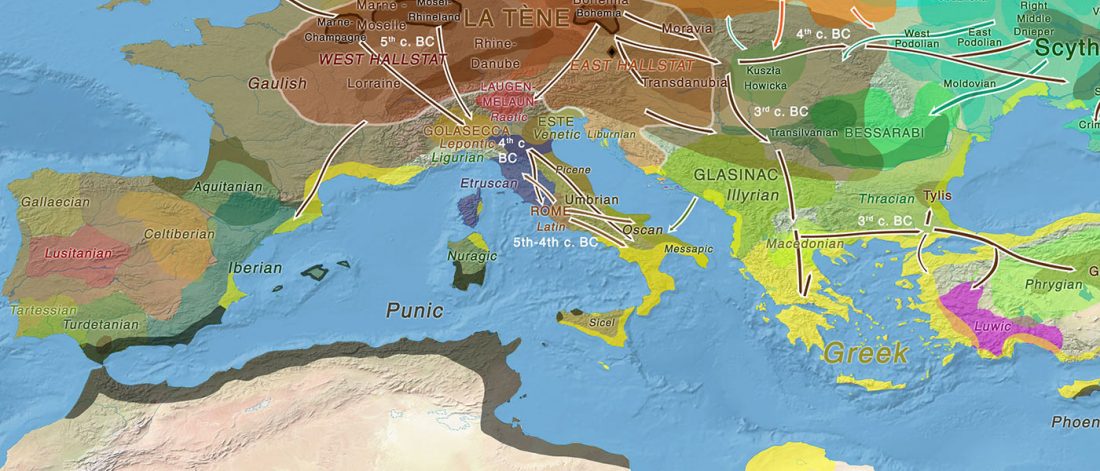
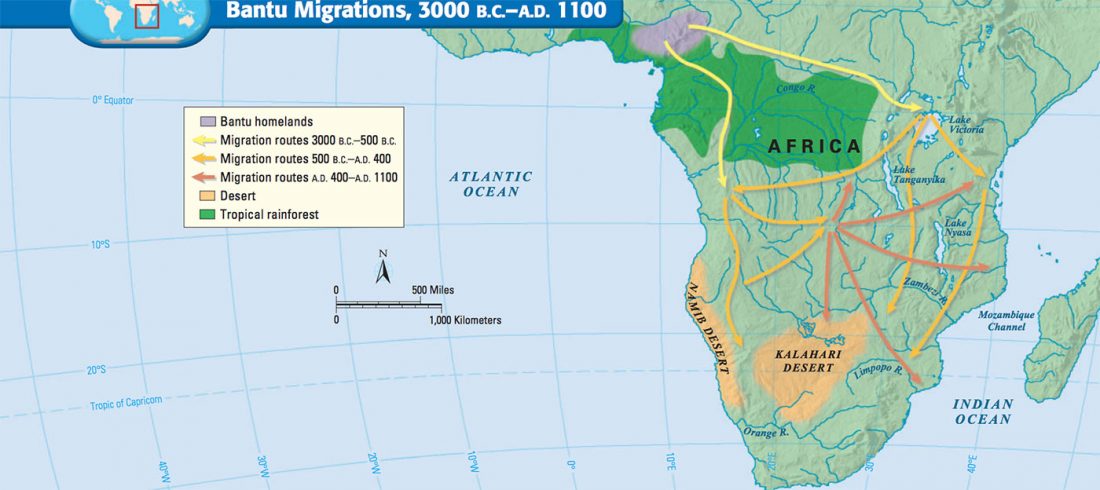
Within months, it will be finally confirmed that both Late Repin offshoots – Early Yamnaya and Afanasievo – spread with clans that were dominated by R1b-L23 patrilineages. Succeeding migration events, likely coupled with internal founder effects under the most successful clans, left Indo-Tocharian-speaking clans as an almost uniform community in terms of Y-chromosome haplogroups, with their most recent common ancestor traceable to the 5th millennium BC.
Before that, it seems that the Indo-Anatolian-speaking Early Khvalynsk community was slightly more diverse. In particular, the success of R1b-V1636 lineages is apparent in the Khvalynsk-Novodanilovka expansion, since it is … Read the rest “Proto-Indo-European kinship system and patrilineality”