Thanks to Joshua Jonathan, I have discovered the paper Massive Migrations? The Impact of Recent aDNA Studies on our View of Third Millennium Europe, by Martin Furholt, European Journal of Archaeology (28 SEP 2017).
Abstract:
New human aDNA studies have once again brought to the forefront the role of mobility and migration in shaping social phenomena in European prehistory, processes that recent theoretical frameworks in archaeology have downplayed as an outdated explanatory notion linked to traditional culture history. While these new genetic data have provided new insights into the population history of prehistoric Europe, they are frequently
…
Read the rest “Massive Migrations? The Impact of Recent aDNA Studies on our View of Third Millennium Europe”
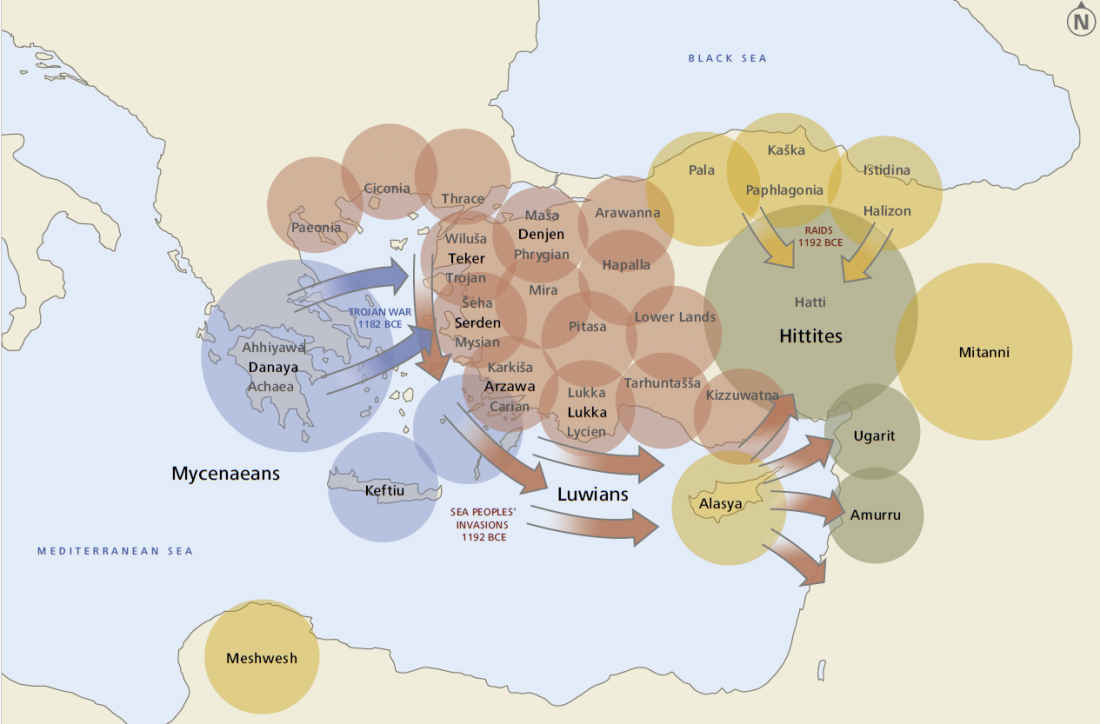
A very interesting monograph on the Luwian Civilization, and its potential connection with Wilusa (Troy) from the end of the third millennium and throughout the Bronze Age: The Luwian Civilization – The Missing Link in the Aegean Bronze Age, by Eberhard Zangger (also available in German: Die luwische Kultur – Das fehlende Element in der Ägäischen Bronzezeit).
Abstract:
Aegean prehistory suffered from a bias when the field was conceived 100 years ago and subsequent research has never questioned the fundamental paradigms of the discipline. As a consequence, only one third of the Aegean coasts have thus far
…
Read the rest “Luwians: the missing link with the Aegean Bronze Age, including Troy and the Sea Peoples”
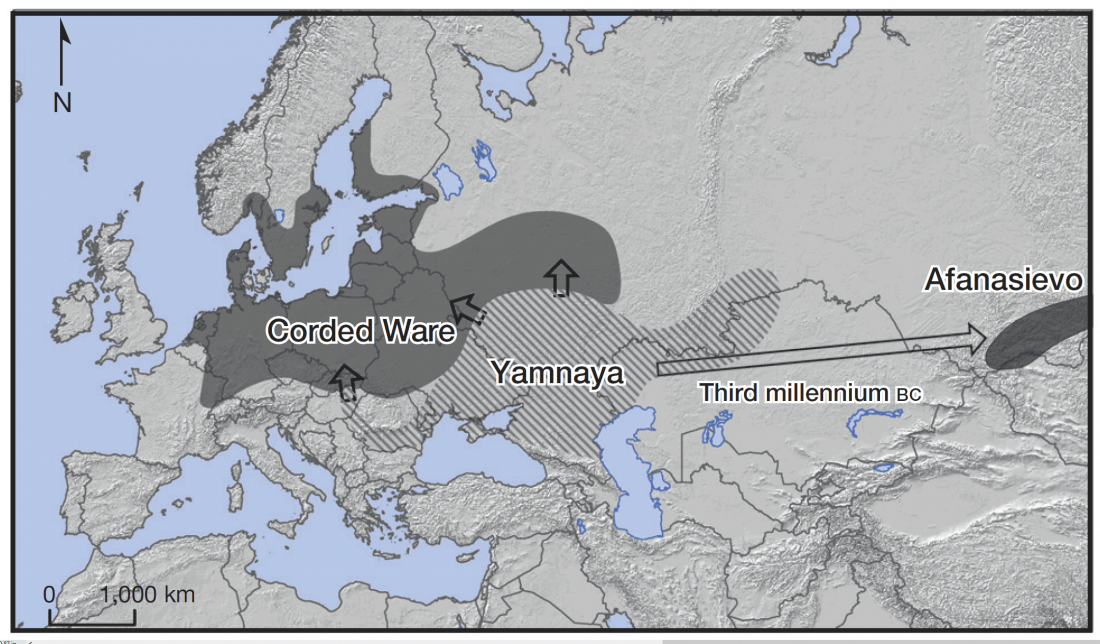
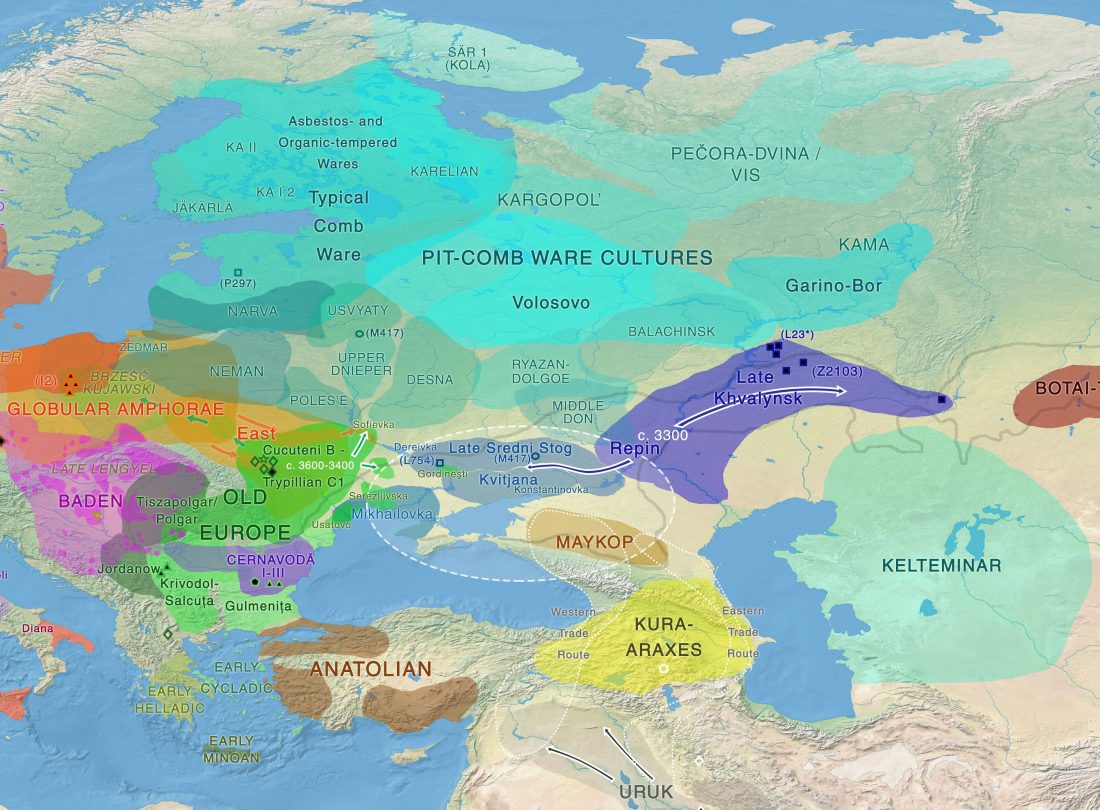
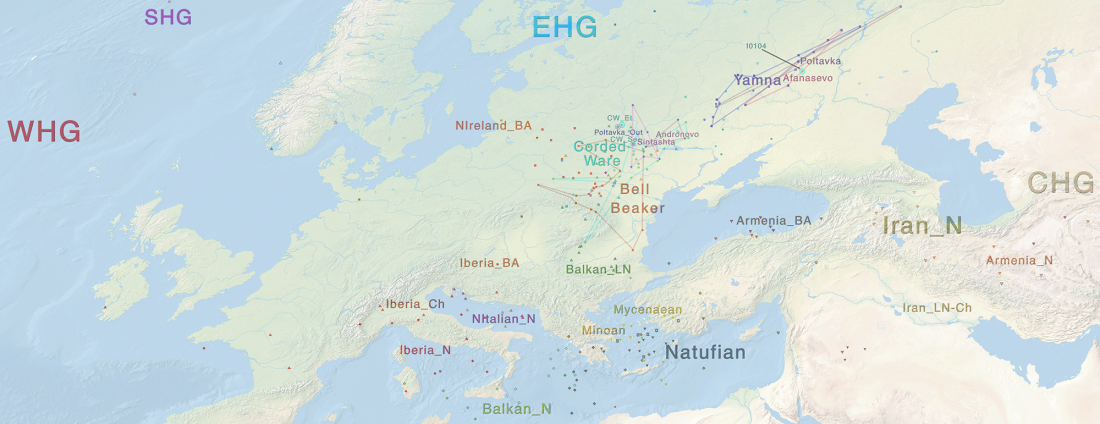
I recently wrote about the Indo-European Corded Ware Theory of Kristian Kristiansen and his workgroup, a sort of “Danish school”, whose aim is to prove a direct, long-lasting interaction between the North Pontic steppe and east European cultures during the Late Neolithic, which supposedly gave rise to a Late Indo-European-speaking Corded Ware culture. That is, a sort of renewed Kurgan model; or, more exactly, Kurgan models, since there is no single one preferred right now.
David Anthony had remained more or less in the background after the controversial assessment of the so-called Yamnaya ancestral component by recent … Read the rest “The new “Indo-European Corded Ware Theory” of David Anthony”
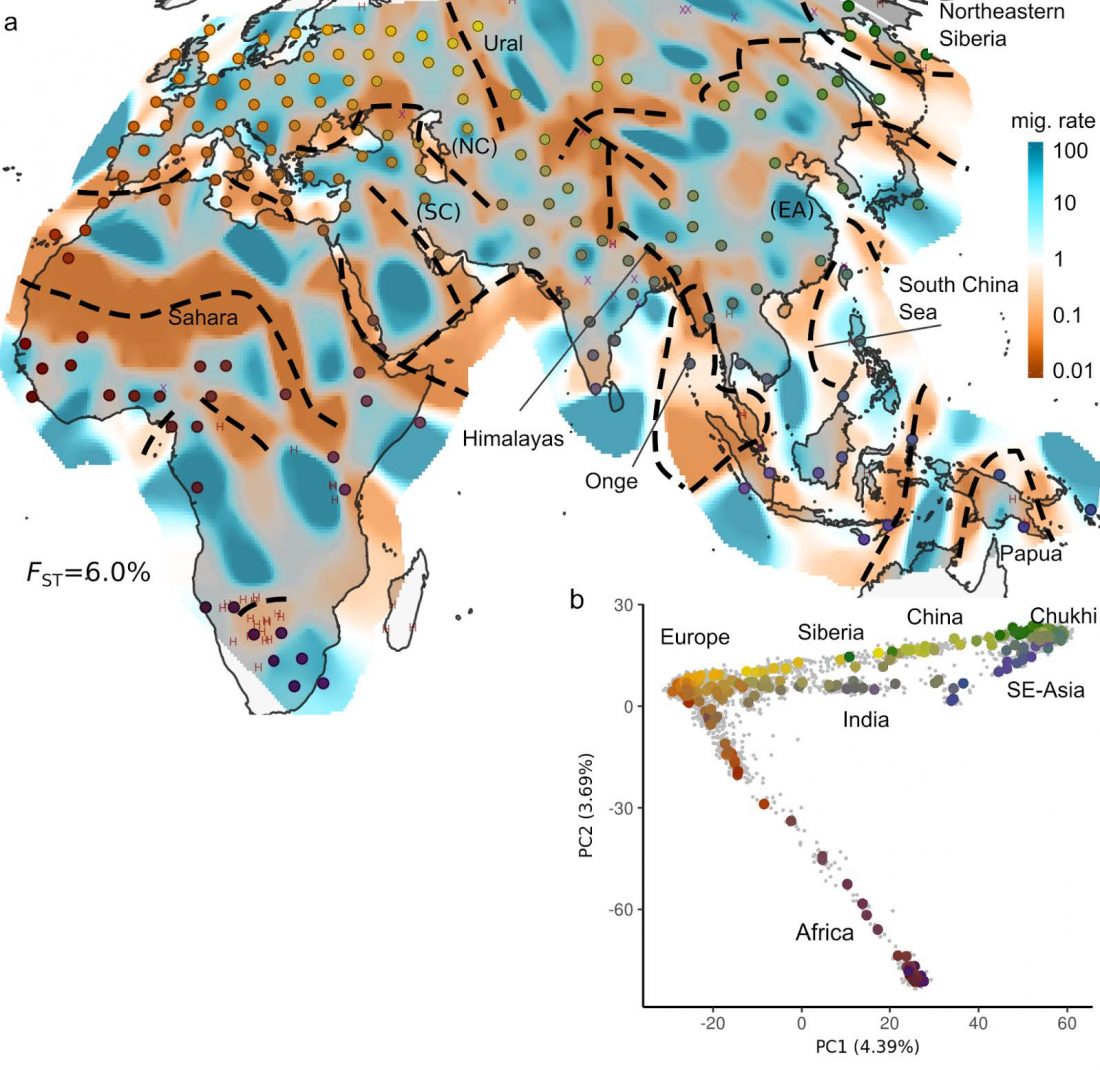
New preprint at BioRxiv, Genetic landscapes reveal how human genetic diversity aligns with geography, by Peter, Petkova, and Novembre (2017).
Abstract:
Summarizing spatial patterns in human genetic diversity to understand population history has been a persistent goal for human geneticists. Here, we use a recently developed spatially explicit method to estimate “effective migration” surfaces to visualize how human genetic diversity is geographically structured (the EEMS method). The resulting surfaces are “rugged”, which indicates the relationship between genetic and geographic distance is heterogenous and distorted as a rule. Most prominently, topographic and marine features regularly align with increased genetic
…
Read the rest “Genetic landscapes showing human genetic diversity aligning with geography”
New preprint paper at BioRxiv, led by a Japanese researcher, with analysis of mtDNA of Trypillians from Verteba Cave, Analysis of ancient human mitochondrial DNA from Verteba Cave, Ukraine: insights into the origins and expansions of the Late Neolithic-Chalcolithic Cututeni-Tripolye Culture, by Wakabayashi et al. (2017).
Abstract:
Background: The Eneolithic (~5,500 yrBP) site of Verteba Cave in Western Ukraine contains the largest collection of human skeletal remains associated with the archaeological Cucuteni-Tripolye Culture. Their subsistence economy is based largely on agro-pastoralism and had some of the largest and most dense settlement sites during the Middle Neolithic in
…
Read the rest “mtDNA haplogroup frequency analysis from Verteba Cave supports a strong cultural frontier between farmers and hunter-gatherers in the North Pontic steppe”
I wrote two days ago in the post anouncing the revised version (October 2017) of the Indo-European demic diffusion model, about dumping the information I had on doing PCA and ADMIXTURE analyses as ‘drafts’, without reviewing them, in the new section of this website called Human Ancestry.
I had some time today to review them, and to correct gross mistakes in the texts, so that they might be more usable now…
I began to work with free datasets to see if I could learn something more about results of recent Genetic research by working with the available … Read the rest “Human ancestry: how to work your own PCA, ADMIXTURE analyses for human evolutionary and genealogical studies”
Last modified: 1st November 2017
AdmixTools
Install admixtools
You can do it with git (you have to have it installed):
sudo apt-get install git
git clone https://github.com/DReichLab/AdmixTools.git
(you can add at the end the folder where you want it downloaded).
There you have the AdmixTools folder.
Another option, if you don’t like git, is wget (or just download the zip directly):
wget https://github.com/DReichLab/AdmixTools/archive/master.zip
unzip master.zip
And there you have the AdmixTools-master folder.
It has to be installed from source, and I had the following packages installed – however, probably just the basic packages (GSL, lapack and openblas, in bold) would … Read the rest “AdmixTools: qpgraph, qp3Pop (f_3 test), qpBound, qpDstat, qpF4ratio, rollof”
Last modified: 1st November 2017
Instructions to obtain a Principal Component Analysis from public datasets
We need a parameter file for smartpca. For example I used the file smartpcaMinSS.par:
genotypename: MinSS.geno
snpname: MinSS.snp
indivname: MinSS.ind
evecoutname: MinSS.pca.evec
evaloutname: MinSS.pca.eval
numoutevec: 10
numoutlieriter: 0
maxpops: 100
poplistname: poplistname.txt
snpweightoutname: MinSS.snpweight
lsqproject: YES
The first three lines use the output of your merged dataset.
The next two lines are the names of the output files, one for eigenvectors and one for eigenvalues.
You probably want to select how many PCs will be output, and (important for ancient populations) you might not want … Read the rest “Principal Component Analysis (PCA) with Eigensoft and R”
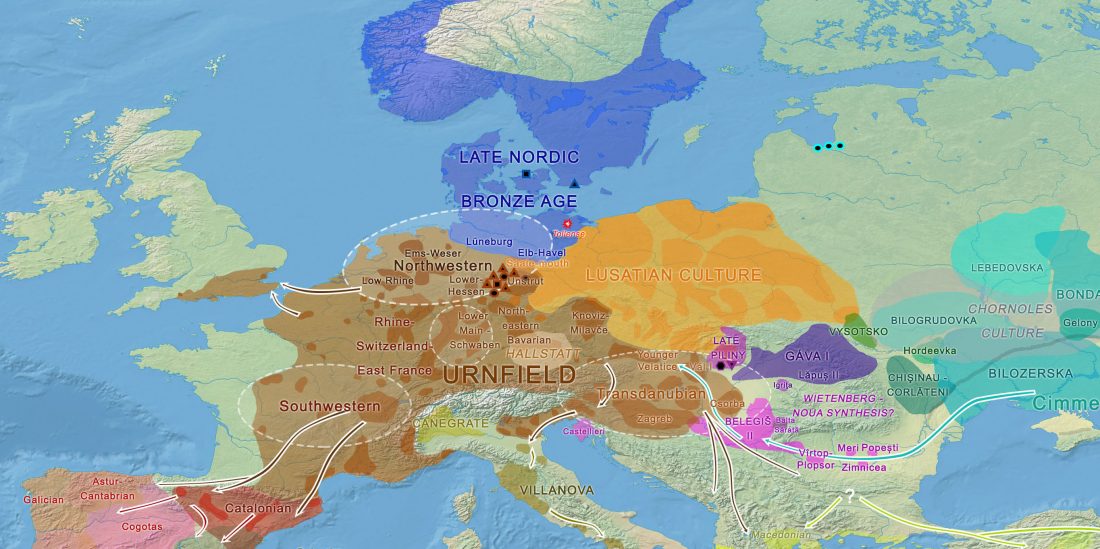
It was reported long ago that genetic studies were being made on remains of a surprisingly big battle that happened in the Tollense valley in north-eastern Germany, at the confluence between Nordic, Tumulus/Urnfield, and Proto-Lusatian/Lusatian territories, ca. 1200 BC.
At least 130 bodies and 5 horses have been identified from the bones found. Taking into account that this is a small percentage of the potential battlefield, around 750 bodies are expected to be buried in the riverbank, so an estimated 4,000-strong army fought there, accounting for one in five participants killed and left on the battlefield.
Body armour, … Read the rest “The Tollense Valley battlefield: the North European ‘Trojan war’ that hints to western Balto-Slavic origins”