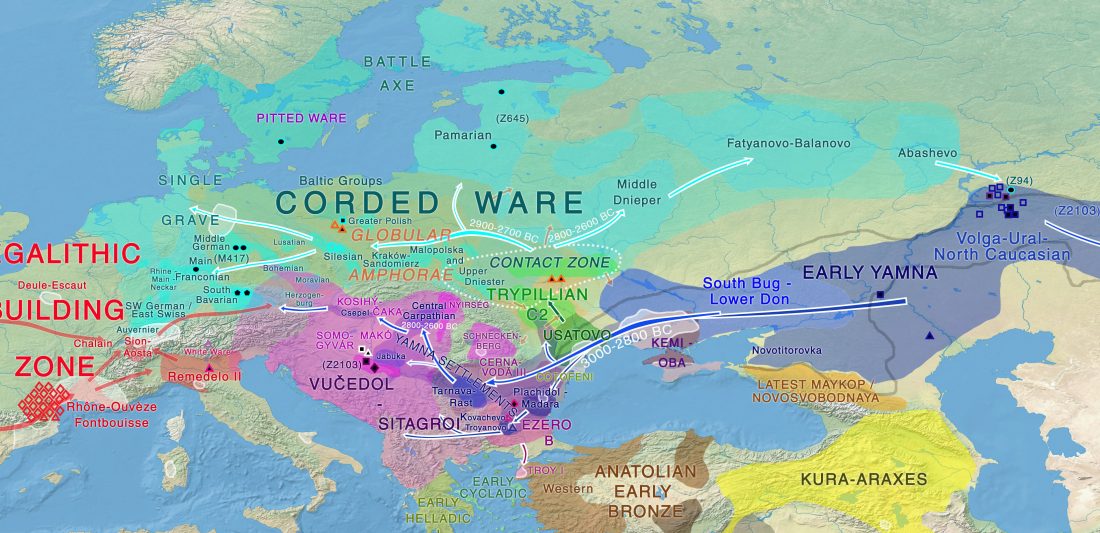
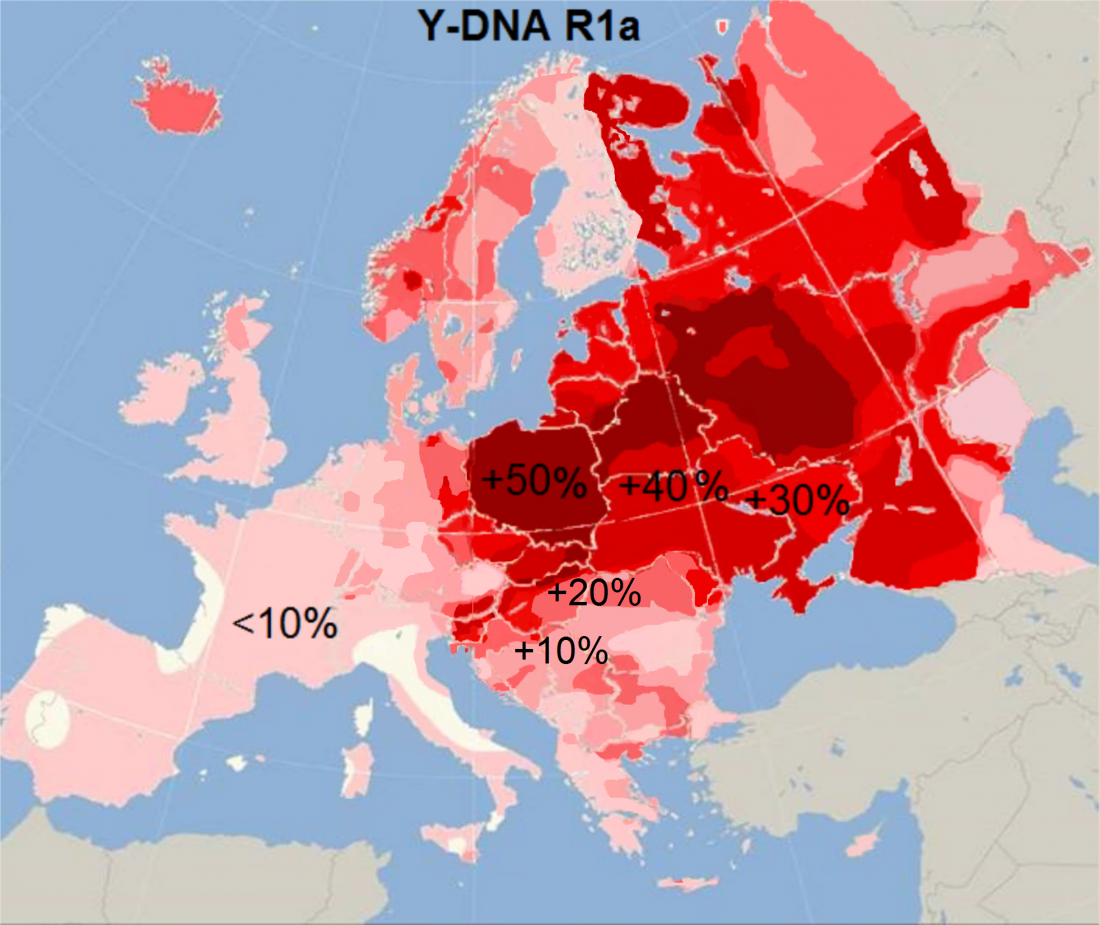
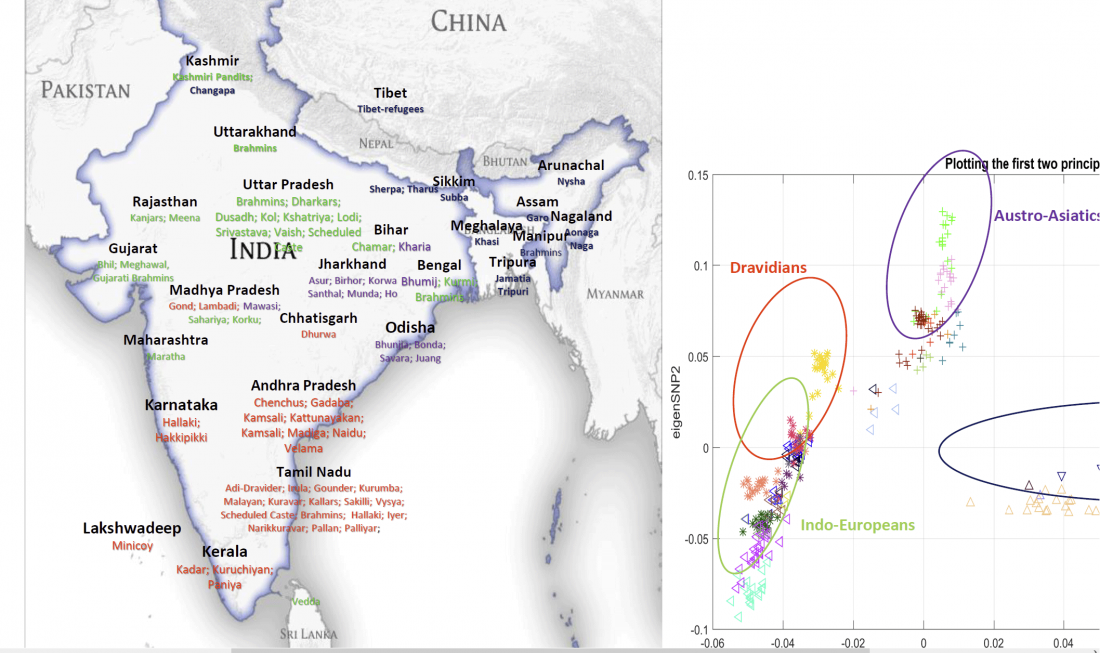
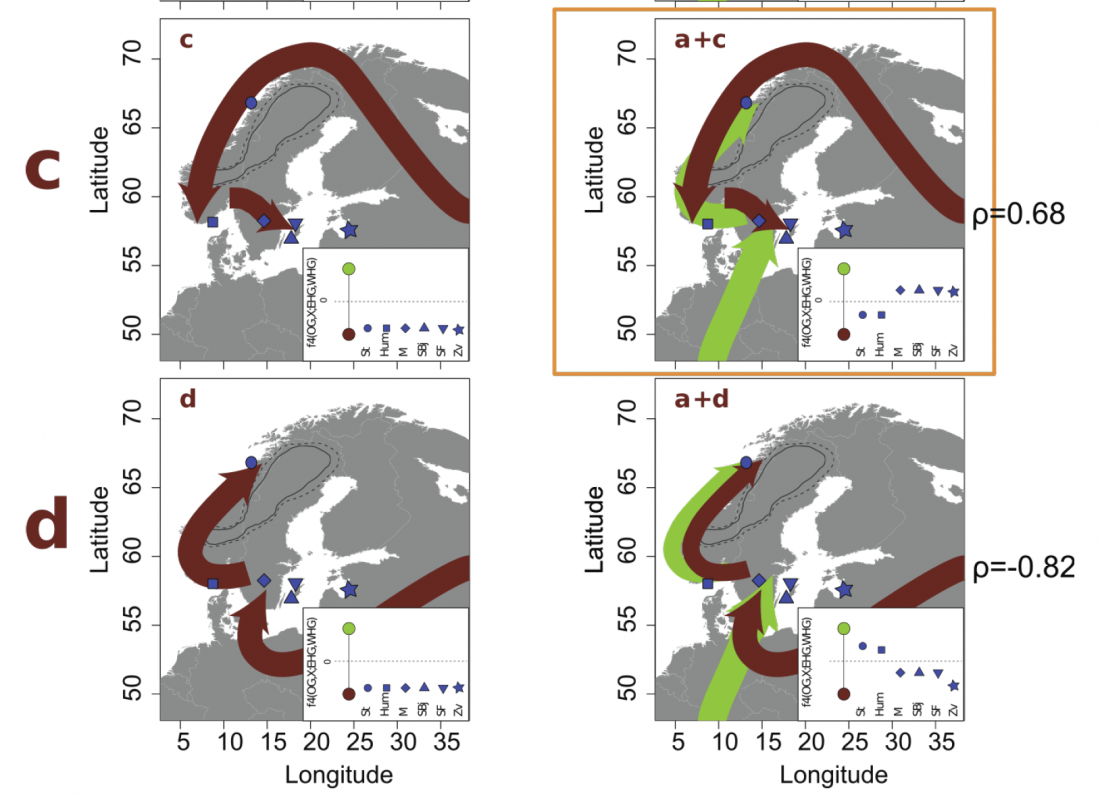
Neolithic and Bronze Age Basque-speaking Iberians resisted invaders from the steppe
Good clickbait, right? I have received reports about this new paper in Google Now the whole weekend, and their descriptions are getting worse each day.
The original title of the article published in PLOS Genetics (already known by its preprint in BioRxiv) was The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods, by Martiniano et al. (2017).
Maybe the title was not attractive enough, so they sent the following summary, entitled “Bronze Age Iberia received fewer Steppe invaders than the rest of Europe” (also in Phys.org. … Read the rest “Neolithic and Bronze Age Basque-speaking Iberians resisted invaders from the steppe”