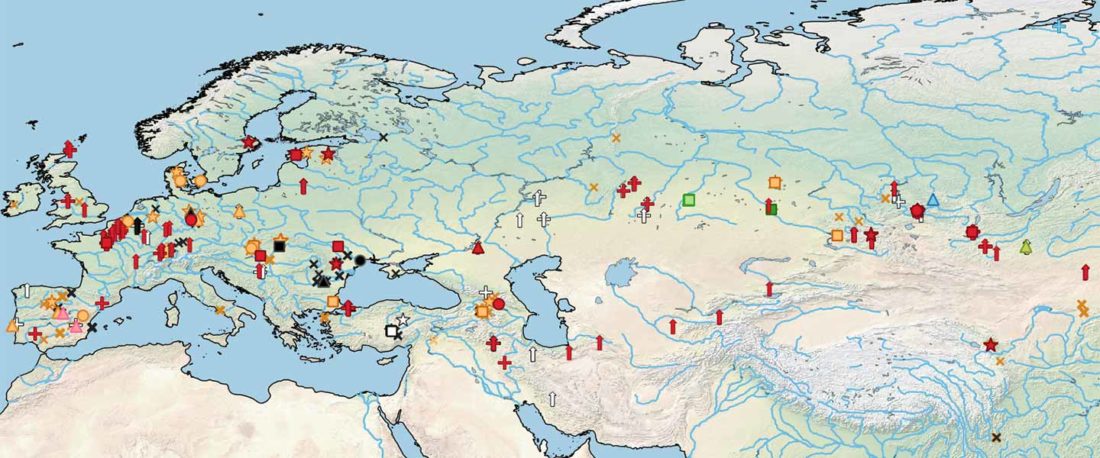
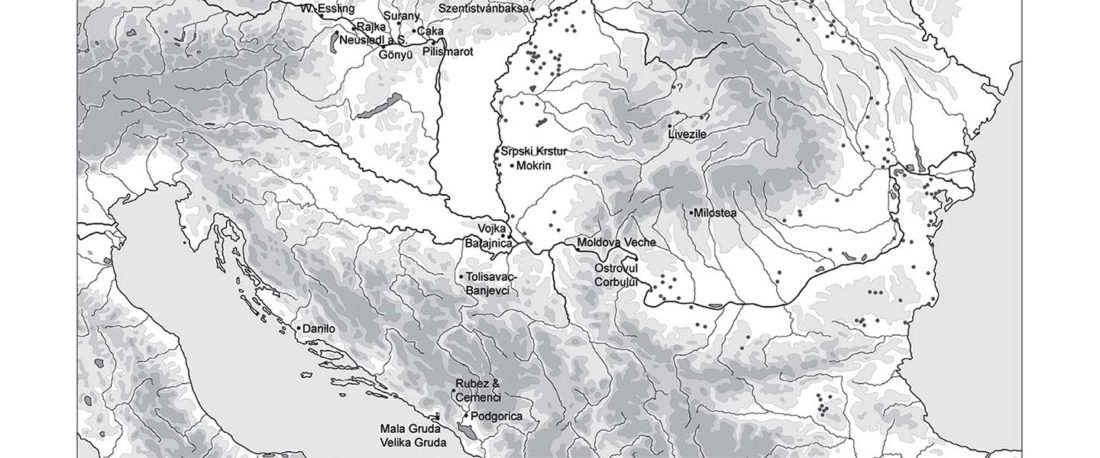
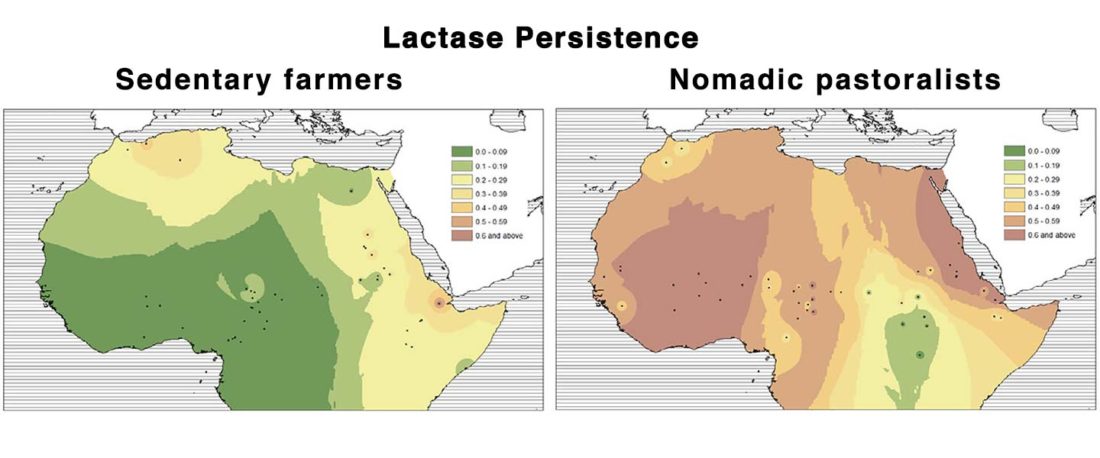
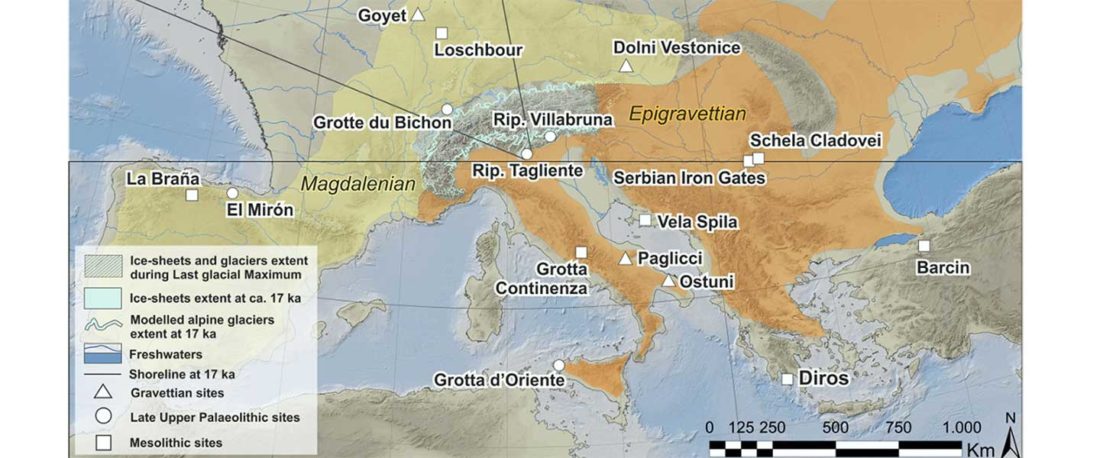
Origin of DOM2 closing in on the Pontic-Caspian steppes
Open access Ancient DNA shows domestic horses were introduced in the southern Caucasus and Anatolia during the Bronze Age by Guimaraes et al. Sci Adv. (2020) Vol. 6, no. 38, eabb0030.
Here is a good summary:
… Read the rest “Origin of DOM2 closing in on the Pontic-Caspian steppes”Our study of ancient equid remains from Anatolia and the southern Caucasus covering ~9000 years of the Holocene analyzed the dynamics over time of mitochondrial lineages and tested the hypothesis that Anatolia was a center of horse domestication. We were able to identify mitotypes characteristic of local Anatolian wild horses, which were regularly exploited in the early and middle Holocene. However, we identified a