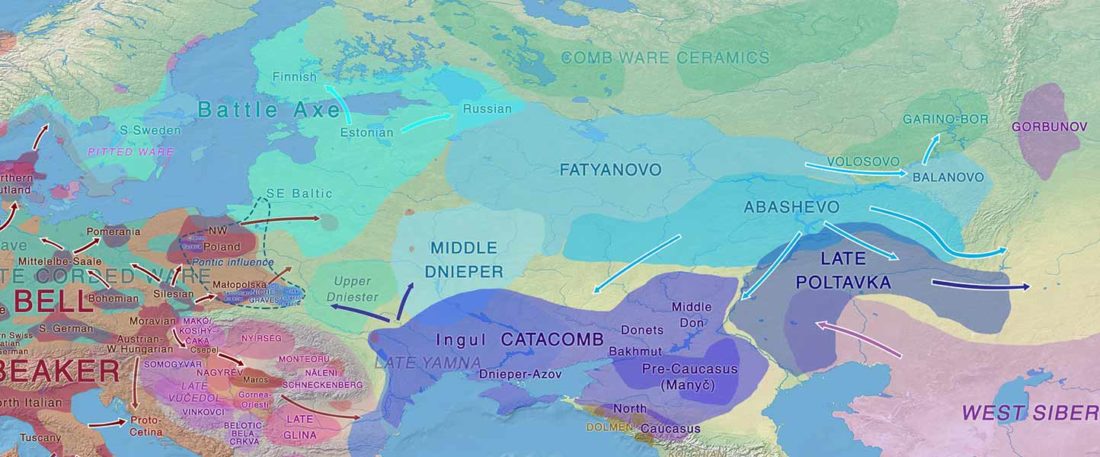
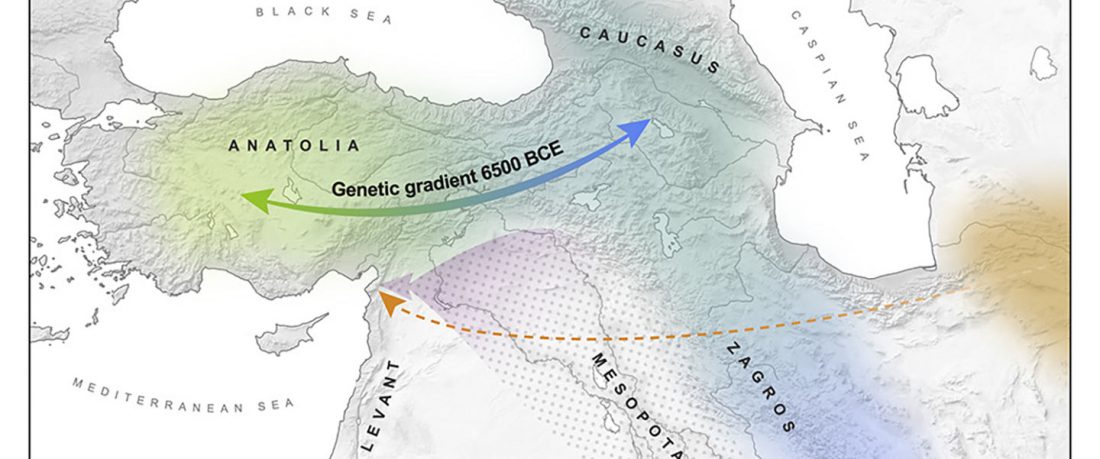
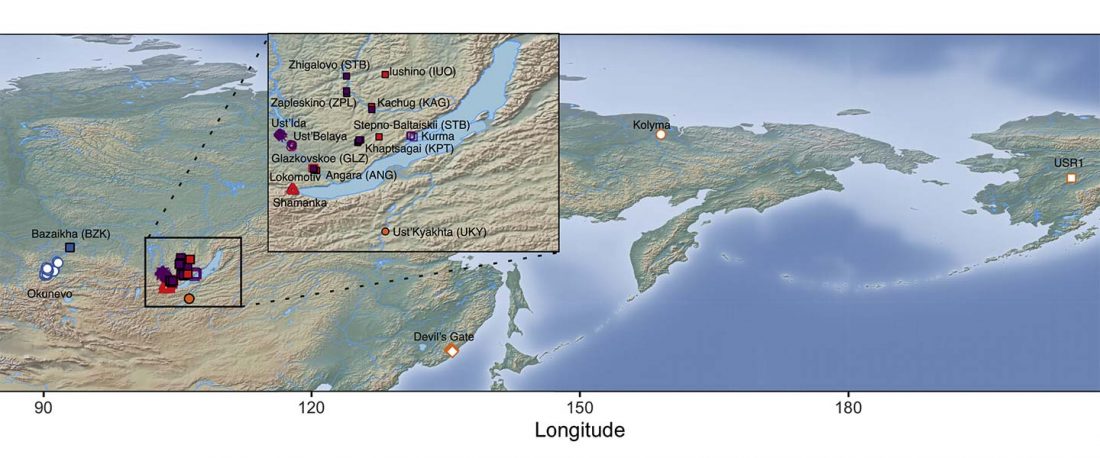
Proto-Uralic Homeland (V): Technology & Trade
This post is part of a draft on palaeolinguistics and the Proto-Uralic homeland. See below for the color code of protoforms.
8. Technology
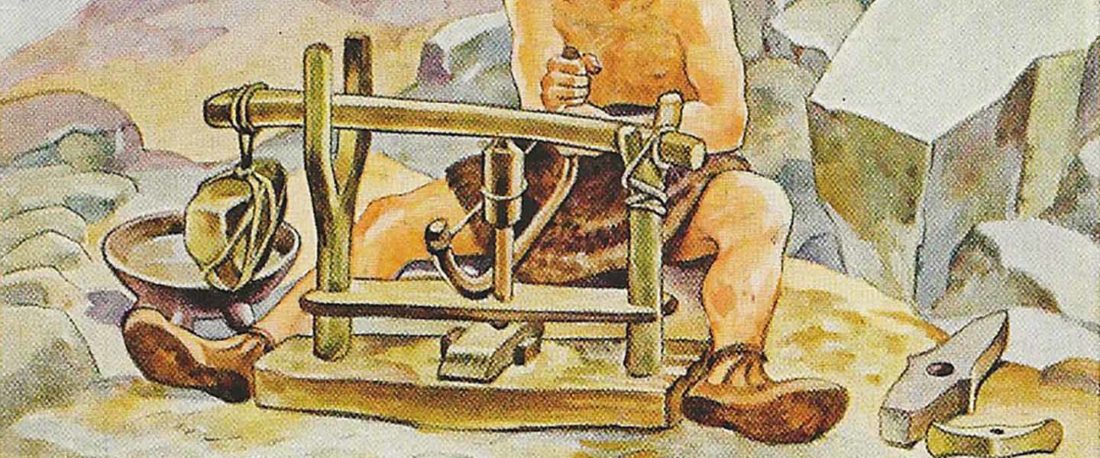
8.1. Pottery
PU (Fi., Ma., Kh., Ms., Hu.) *pata ‘pot’ (UEW Nº 710) has a striking resemblance with NWIE *pod-óm, cf. PGmc. *fatą ‘vat, vessel’, Lith. púodas ‘pot’ (Kroonen 2013: 131; Dérksen 2015: 372). However, a Pre-PGmc. origin of a PFU stem seems unlikely – based on the lack of any other case with such a large distribution. Assuming that an unattested PIIr. **padá- underlies the PU form (cf. Parpola … Read the rest “Proto-Uralic Homeland (V): Technology & Trade”