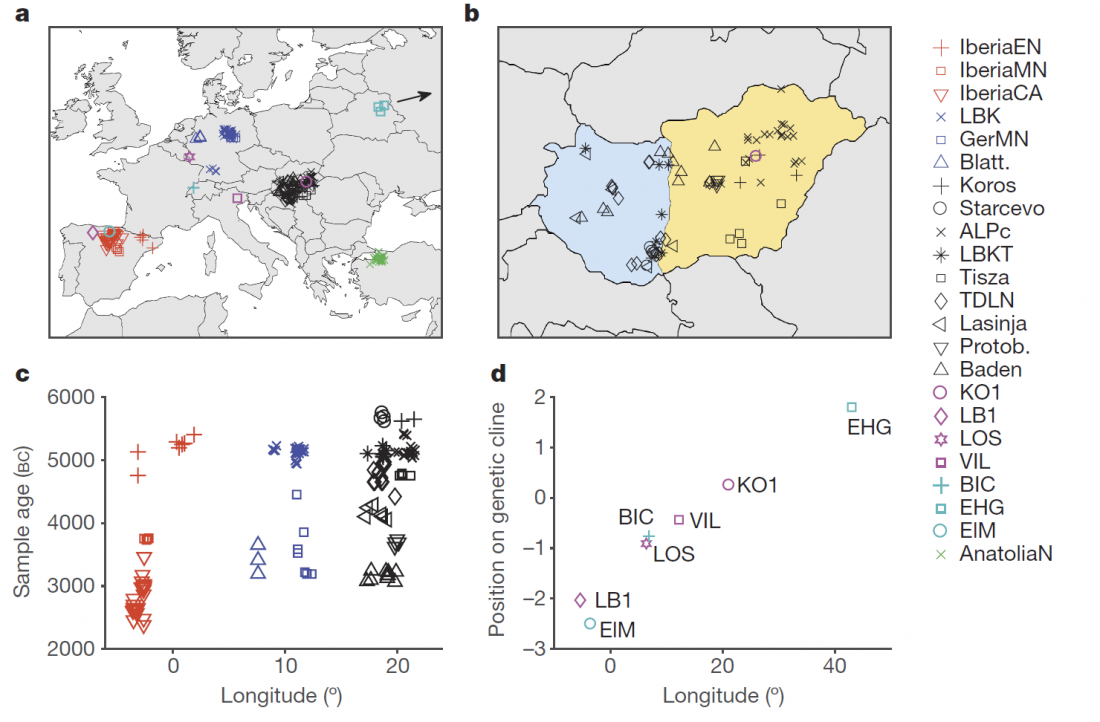
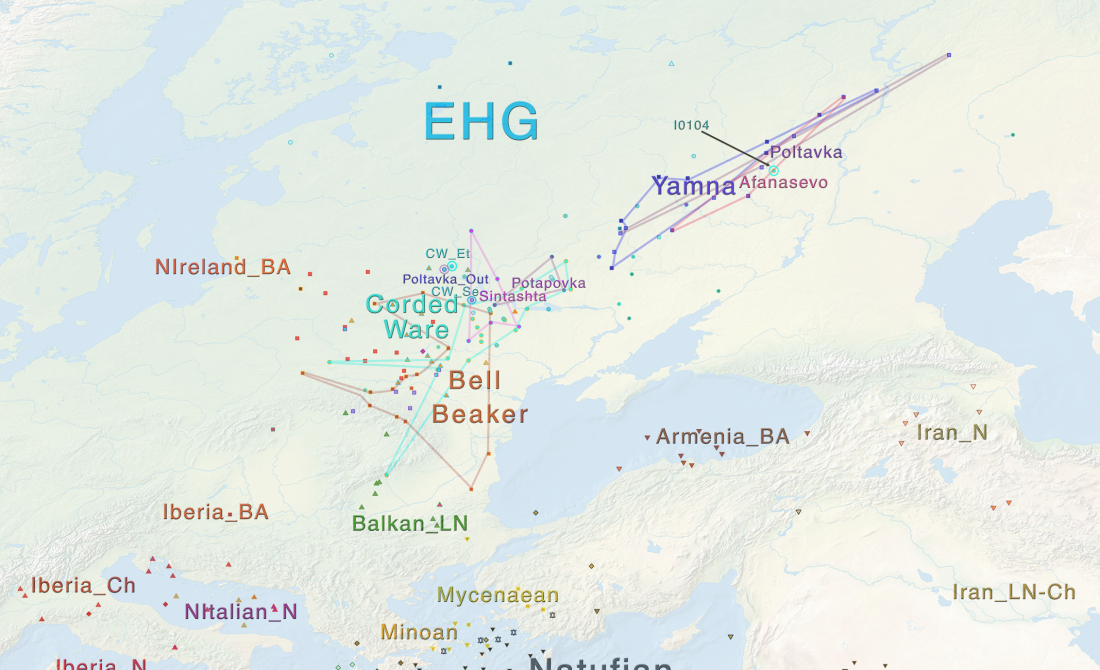
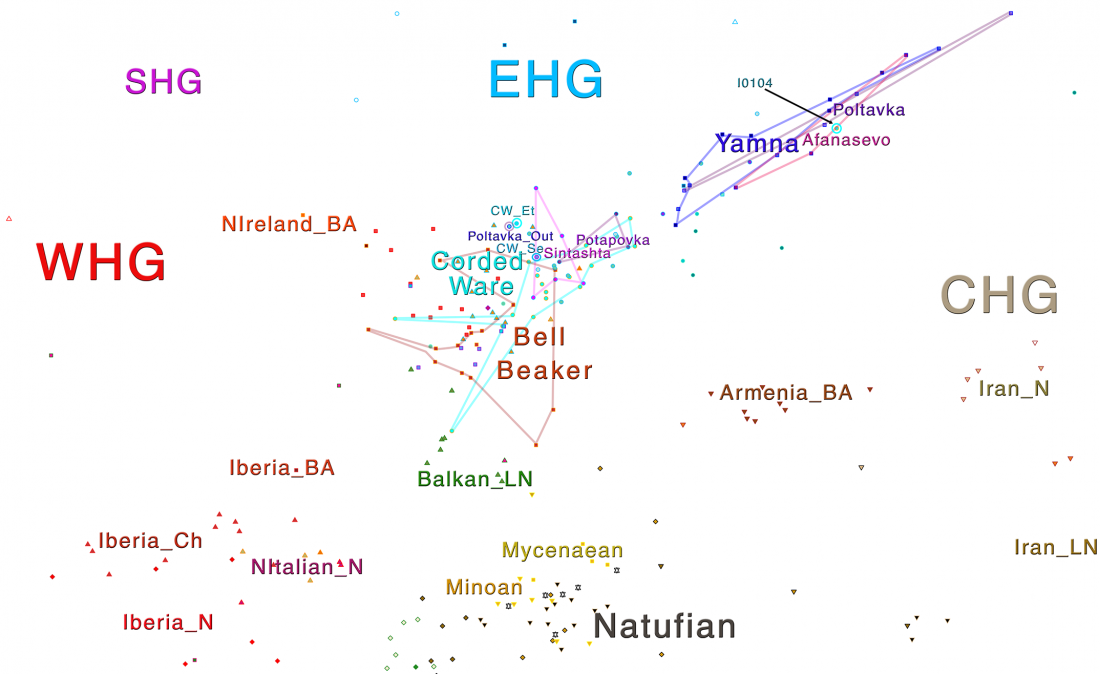
The definitive publication of a BioRxiv preprint article, in Nature: Parallel palaeogenomic transects reveal complex genetic history of early European farmers, by Lipson et al. (2017).
The dataset with all new samples is available at the Reich Lab’s website. You can try my drafts on how to do your own PCA and ADMIXTURE analysis with some of their new datasets.
Abstract:
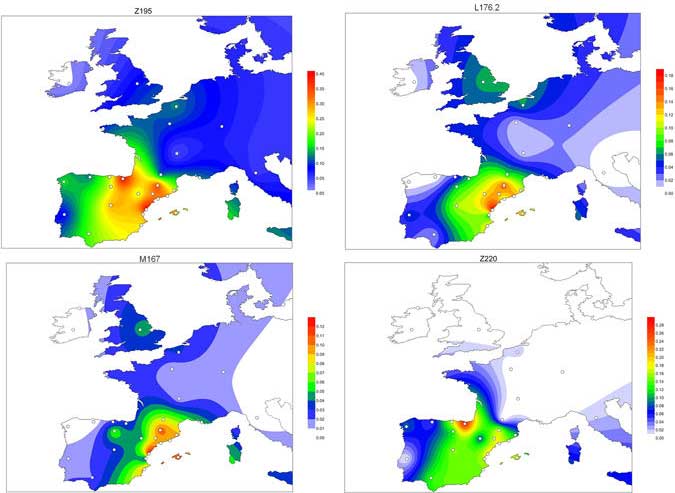
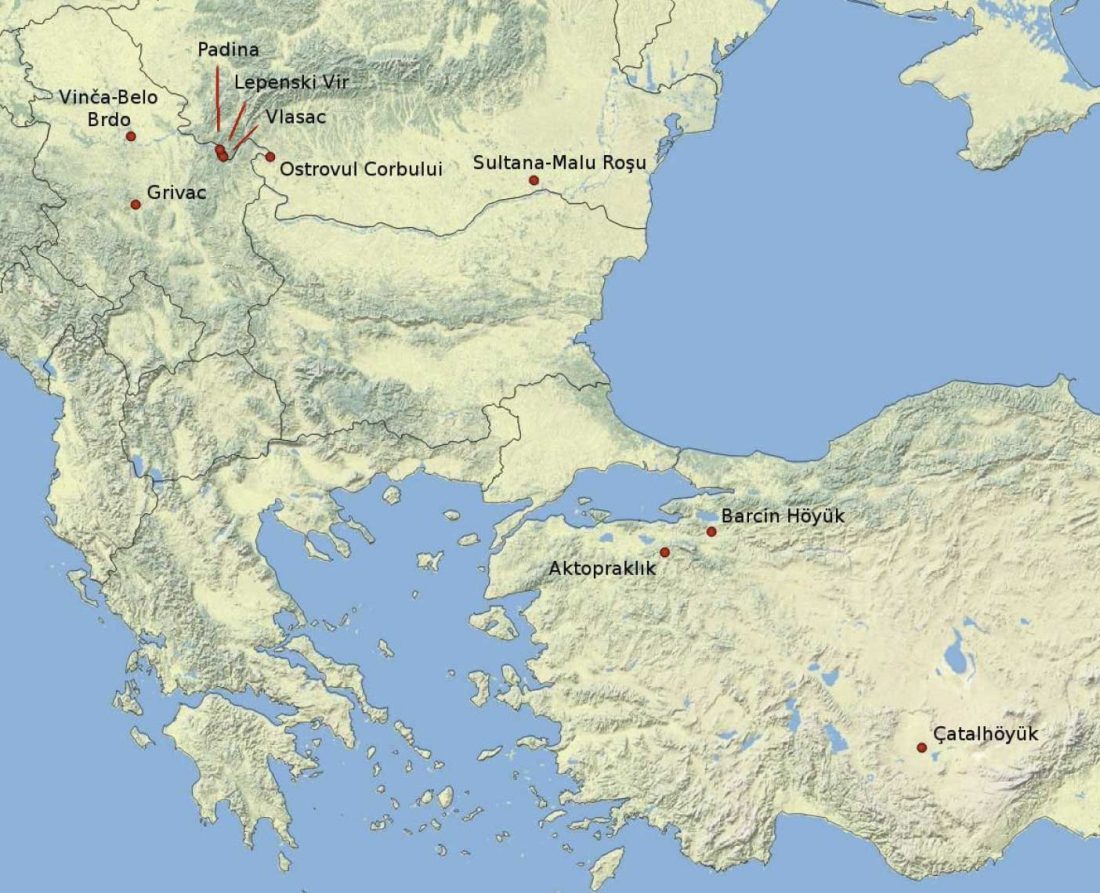
… Read the rest “Before steppe ancestry: Europe’s genetic diversity shaped mainly by local processes, with varied sources and proportions of hunter-gatherer ancestry”Ancient DNA studies have established that Neolithic European populations were descended from Anatolian migrants who received a limited amount of admixture from resident hunter-gatherers. Many open questions remain, however, about the spatial and temporal dynamics of