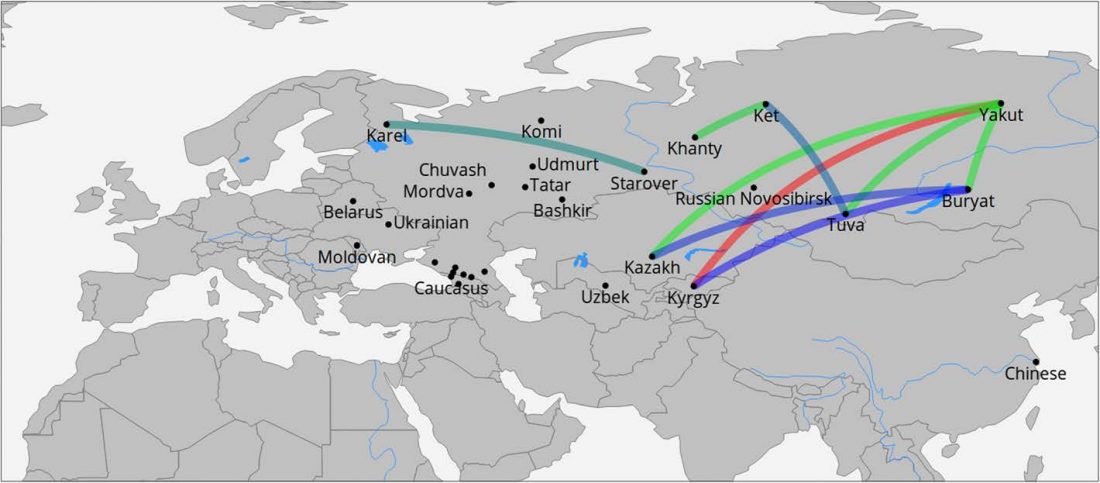
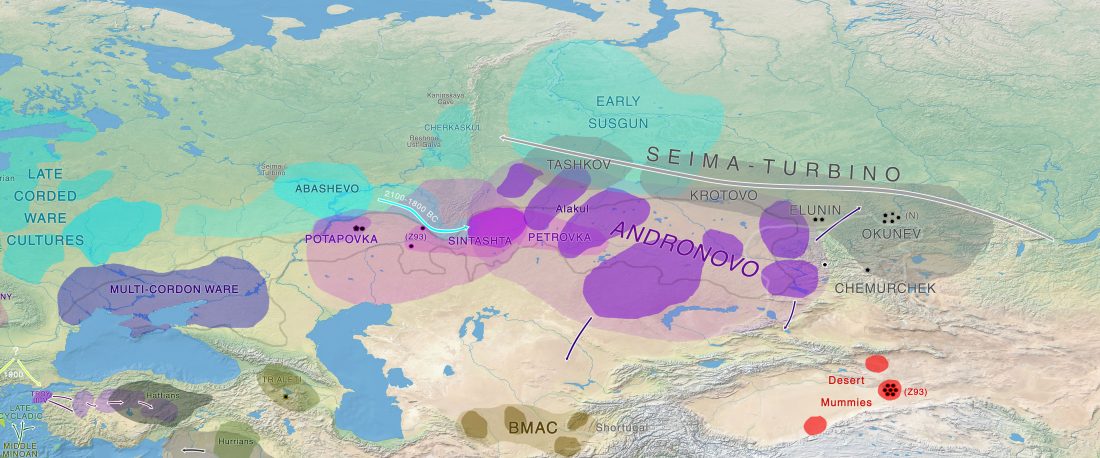
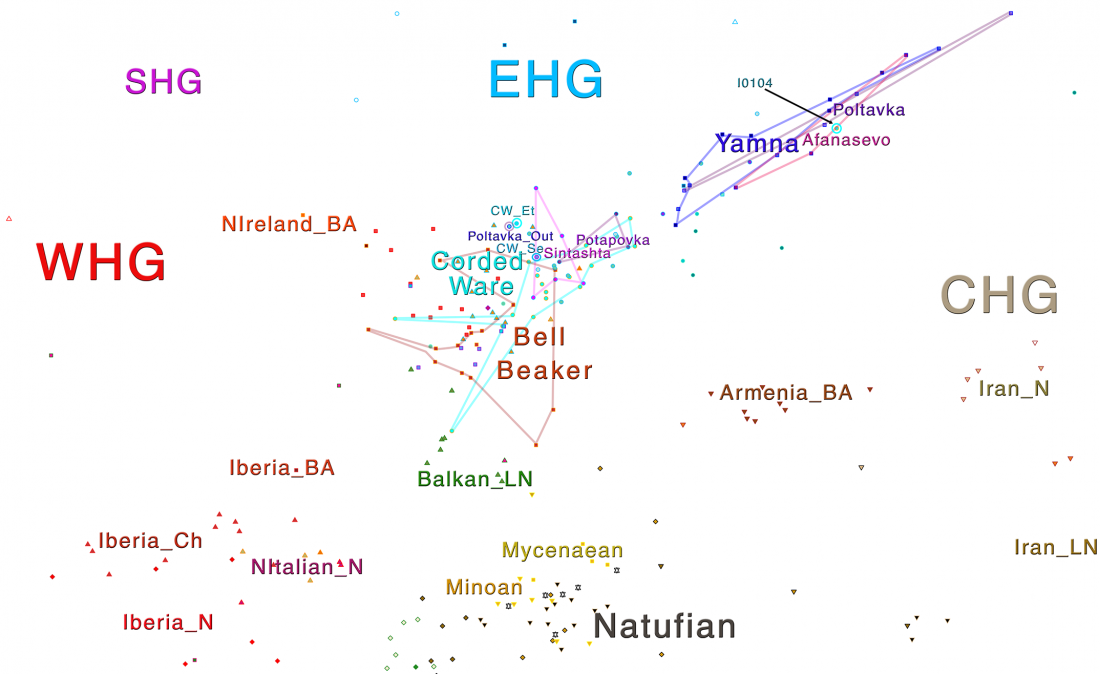
Open Access article on modern populations (including ancient samples), Between Lake Baikal and the Baltic Sea: genomic history of the gateway to Europe, by Triska et al., BMC Genetics 18(Suppl 1):110, 2017.
Abstract:
… Read the rest “Genomic history of Northern Eurasians includes East-West and North-South gradients”Background
The history of human populations occupying the plains and mountain ridges separating Europe from Asia has been eventful, as these natural obstacles were crossed westward by multiple waves of Turkic and Uralic-speaking migrants as well as eastward by Europeans. Unfortunately, the material records of history of this region are not dense enough to reconstruct details of population history. These considerations stimulate growing interest to obtain