New paper (behind paywall) The female ancestor’s tale: Long‐term matrilineal continuity in a nonisolated region of Tuscany, by Leonardi et al. Am J Phys Anthr (2018).
EDIT (10 SEP 2018): The main author has shared an open access link to read the PDF.
Interesting excerpts:
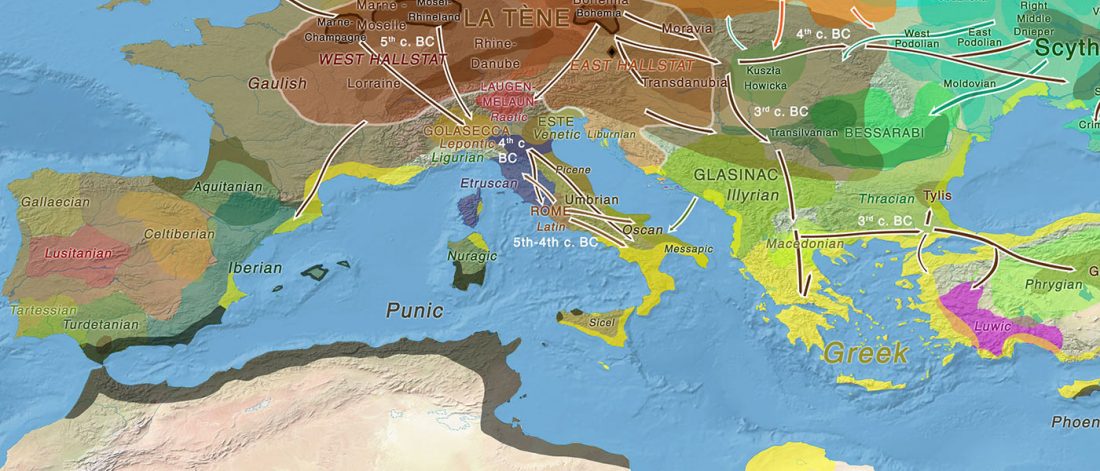
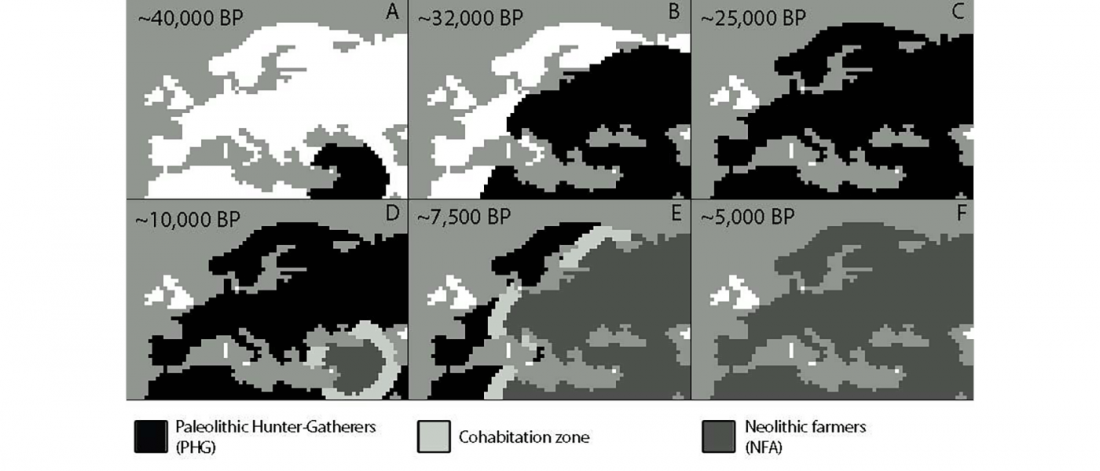
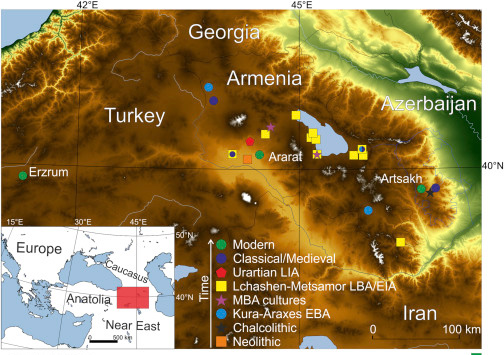
… Read the rest “Long-term matrilineal continuity in a nonisolated region of Tuscany”Here we analyze North-western Tuscany, a region that was a corridor of exchanges between Central Italy and the Western Mediterranean coast.
We newly obtained mitochondrial HVRI sequences from 28 individuals, and after gathering published data, we collected genetic information for 119 individuals from the region. Those span five periods during the last 5,000